Abstract
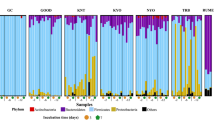
This study evaluated the effects of nitrogen source composition and monensin on the populations of proteolytic and amino acid-fermenting bacteria using in vitro enrichment culture. The experiment was designed with a 2 × 2 factorial arrangement: two nitrogen sources, casein (Cas) and tryptone (Try), and two levels of monensin, 0 (C) and 5 µmol/L (M), resulting in four treatments: Cas-C, Cas-M, Try-C, and Try-M. Ruminal fluid collected from three cannulated Holstein dairy cows was used as the inoculum. Each treatment culture was consecutively transferred six times after 24 h of incubation. The results showed that ammonia concentration was lower in Cas than in Try, and it was reduced by monensin addition. In the 6th transfer enrichment cultures, the 16S rRNA gene copy numbers of total bacteria were reduced by monensin but was unaffected by nitrogen sources. Principal component analysis showed that the bacterial communities differed among the treatments. At the genus level, Peptostreptococcus accounted for as much as 41% of the total bacteria in Try-C, but it made up less than 0.02% in the other three treatments. A Pearson correlation analysis showed that the relative abundance of Peptostreptococcus was positively correlated with ammonia concentration. Overall, the results suggest that nitrogen source composition and monensin can affect ruminal ammonia production by modulating the ruminal proteolytic bacterial communities, and some hyper-ammonia-producing bacteria of the genus Peptostreptococcus may be among the main culprits contributing to the high ammonia concentration in the rumen.





Similar content being viewed by others
References
Calsamiglia S, Ferret A, Reynolds CK, Kristensen NB, van Vuuren AM (2010) Strategies for optimizing nitrogen use by ruminants. Animal 4(7):1184–1196. https://doi.org/10.1017/S1751731110000911
Kohn RA, Dinneen MM, Russek-Cohen E (2005) Using blood urea nitrogen to predict nitrogen excretion and efficiency of nitrogen utilization in cattle, sheep, goats, horses, pigs, and rats. J Anim Sci 83(4):879–889. https://doi.org/10.2527/2005.834879x
Tamminga S (1992) Nutrition management of dairy-cows as a contribution to pollution-control. J Dairy Sci 75(1):345–357
Tamminga S (1996) A review on environmental impacts of nutritional strategies in ruminants. J Anim Sci 74(12):3112–3124
Walker ND, Newbold CJ, Wallace RJ (2005) Nitrogen metabolism in the rumen. In: Pfeffer E, Hristov A (eds) Nitrogen and phosphorus nutrition of cattle. CABI Publishing, Cambridge, pp 71–115
Bach A, Calsamiglia S, Stern MD (2005) Nitrogen metabolism in the rumen. J Dairy Sci 88(13):E9–E21
Wallace RJ (1996) Ruminal microbial metabolism of peptides and amino acids. J Nutr 126(4):S1326–S1334
Paster BJ, Russell JB, Yang CMJ, Chow JM, Woese CR, Tanner R (1993) Phylogeny of the ammonia-producing ruminal bacteria Peptostreptococcus anaerobius, Clostridium sticklandii, and Clostridium aminophilum sp. nov. Int J Syst Bacteriol 43(1):107–110. https://doi.org/10.1099/00207713-43-1-107
Attwood GT, Klieve AV, Ouwerkerk D, Patel BKC (1998) Ammonia-hyperproducing bacteria from New Zealand ruminants. Appl Environ Microb 64(5):1796–1804
McSweeney CS, Palmer B, Bunch R, Krause DO (1999) Isolation and characterization of proteolytic ruminal bacteria from sheep and goats fed the tannin-containing shrub legume Calliandra calothyrsus. Appl Environ Microb 65(7):3075–3083
Eschenlauer SCP, McKain N, Walker ND, McEwan NR, Newbold CJ, Wallace RJ (2002) Ammonia production by ruminal microorganisms and enumeration, isolation, and characterization of bacteria capable of growth on peptides and amino acids from the sheep rumen. Appl Environ Microb 68(10):4925–4931. https://doi.org/10.1128/aem.68.10.4925-4931.2002
Wallace RJ, McKain N, McEwan NR, Miyagawa E, Chaudhary LC, King TP, Walker ND, Apajalahti JHA, Newbold CJ (2003) Eubacterium pyruvativorans sp. nov., a novel non-saccharolytic anaerobe from the rumen that ferments pyruvate and amino acids, forms caproate and utilizes acetate and propionate. Int J Syst Evol Micr 53:965–970. https://doi.org/10.1099/ijs.0.02110-0
Flythe MD, Andries K (2009) The effects of monensin on amino acid catabolizing bacteria isolated from the Boer goat rumen. Small Ruminant Res 81(2–3):178–181. https://doi.org/10.1016/j.smallrumres.2008.12.004
Bento CBP, de Azevedo AC, Detmann E, Mantovani HC (2015) Biochemical and genetic diversity of carbohydrate-fermenting and obligate amino acid-fermenting hyper-ammonia-producing bacteria from Nellore steers fed tropical forages and supplemented with casein. BMC Microbiol 15. https://doi.org/10.1186/s12866-015-0369-9
Chen GJ, Russell JB (1988) Fermentation of peptides and amino-acids by a monensin-sensitive ruminal Peptostreptococcus. Appl Environ Microb 54(11):2742–2749
Chaney AL, Marbach EP (1962) Modified reagents for determination of urea and ammonia. Clin Chem 8:130–132
Dai ZL, Zhang J, Wu GY, Zhu WY (2010) Utilization of amino acids by bacteria from the pig small intestine. Amino Acids 39(5):1201–1215. https://doi.org/10.1007/s00726-010-0556-9
Denman SE, McSweeney CS (2006) Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol 58(3):572–582. https://doi.org/10.1111/j.1574-6941.2006.00190.x
Devkota S, Wang YW, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB (2012) Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10(−/−) mice. Nature 487(7405):104–108. https://doi.org/10.1038/nature11225
Patra AK, Yu Z (2014) Effects of vanillin, quillaja saponin, and essential oils on in vitro fermentation and protein-degrading microorganisms of the rumen. Appl Microbiol Biotechnol 98(2):897–905. https://doi.org/10.1007/s00253-013-4930-x
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Mao S, Zhang M, Liu J, Zhu W (2015) Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci Rep 5:16116. https://doi.org/10.1038/srep16116
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. https://doi.org/10.1093/bioinformatics/btq461
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 72(7):5069–5072. https://doi.org/10.1128/Aem.03006-05
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26(2):266–267. https://doi.org/10.1093/bioinformatics/btp636
Price MN, Dehal PS, Arkin AP (2009) FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26(7):1641–1650. https://doi.org/10.1093/molbev/msp077
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 73(16):5261–5267. https://doi.org/10.1128/AEM.00062-07
Wallace RJ, Mckain N (1991) A survey of peptidase activity in rumen bacteria. J Gen Microbiol 137:2259–2264
Howard BH, Hungate RE (1976) Desulfovibrio of the sheep rumen. Appl Environ Microbiol 32(4):598–602
Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR (2006) Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res 4(1):9–14. https://doi.org/10.1158/1541-7786.MCR-05-0126
Yang G (2011) Hydrogen sulfide in cell survival: a double-edged sword. Expert Rev Clin Pharmacol 4(1):33–47. https://doi.org/10.1586/ecp.10.131
Song Y, Malmuthuge N, Steele MA, Guan LL (2018) Shift of hindgut microbiota and microbial short chain fatty acids profiles in dairy calves from birth to pre-weaning. FEMS Microbiol Ecol 94(3):fix179. https://doi.org/10.1093/femsec/fix179
Anderson RC, Flythe MD, Krueger NA, Callaway TR, Edrington TS, Harvey RB, Nisbet DJ (2010) Decreased competiveness of the foodborne pathogen Campylobacter jejuni during co-culture with the hyper-ammonia producing anaerobe Clostridium aminophilum. Folia Microbiol 55(4):309–311. https://doi.org/10.1007/s12223-010-0046-1
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Award No.: 31402101) and Natural Science Foundation of Jiangsu Province (Award No.: BK20140696). A scholarship from China Scholarship Council partially supported J. S. tenure at The Ohio State University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shen, J., Yu, Z. & Zhu, W. Insights into the Populations of Proteolytic and Amino Acid-Fermenting Bacteria from Microbiota Analysis Using In Vitro Enrichment Cultures. Curr Microbiol 75, 1543–1550 (2018). https://doi.org/10.1007/s00284-018-1558-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1558-1