Abstract
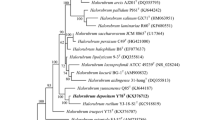
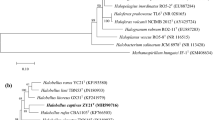
A non-motile, spherical or oval extremely halophilic archaeon, strain Y69T, was isolated from a brine of the Yunnan salt mine, China. Colonies on JCM 168 agar plate were round (1–2 mm in diameter), moist, and orange-pigmented. Phylogenetic analysis of the almost-complete 16S rRNA gene sequence showed that the isolate belonged to the species of the genus Halorubrum, with a close relationship to Halorubrum aidingense 31-hongT (98.5%), Halorubrum lacusprofundi ATCC 49239T (98.2%), and Halorubrum kocurii BG-1T (98.0%). The major polar lipids of strain Y69T were phosphatidylglycerol phosphate methyl ester, phosphatidylglycerol sulfate and a sulfated diglycosyl diether. Strain Y69T grew in 15–30% (w/v) NaCl. The temperature and pH ranges for growth were 25–50 °C and 6.5–9.0, respectively. Optimal growth occurred at 20% (w/v) NaCl, 42 °C, and pH 8.0. Mg2+ was required for growth. The DNA G+C content was determined to be 65.1 mol% by the thermal denaturation method. DNA–DNA hybridization values between strain Y69T and the closely related species were lower than 70%. Based on the data presented in this study, strain Y69T represents a novel species for which the name Halorubrum salsamenti sp. nov. is proposed. The type of the strain is Y69T (=CGMCC 1.15455T = JCM 31270T).


Similar content being viewed by others
References
Bonelo G, Ventosa A, Megias M, Ruiz-Berraquero F (1984) The sensitivity of halobacteria to antibiotics. FEMS Microbiol Lett 21:341–345
Boucher Y, Douady CJ, Sharma AK, Kamekura M, Doolittle WF (2004) Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J Bacteriol 186:3980–3990
Çınar S, Mutlu MB (2016) Comparative analysis of prokaryotic diversity in solar salterns in eastern Anatolia (Turkey). Extremophiles 20:589–601
Corral P, de la Haba RR, Sánchez-Porro C, Amoozegar MA, Papke RT, Ventosa A (2015) Halorubrum persicum sp. nov., an extremely halophilic archaeon isolated from sediment of a hypersaline lake. Int J Syst Evol Microbiol 65:1770–17785
Cui HL, Lin ZY, Dong Y, Zhou PJ, Liu SJ (2007) Halorubrum litoreum sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 57:2204–2206
Cui HL, Tohty D, Zhou PJ, Liu SJ (2006) Halorubrum lipolyticum sp. nov. and Halorubrum aidingense sp. nov., isolated from two salt lakes in Xin-Jiang, China. Int J Syst Evol Microbiol 56:1631–1634
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
DeMaere MZ, Williams TJ, Allen MA, Brown MV, Gibson JA, Rich J, Lauro FM, Dyall-Smith M, Davenport KW, Woyke T, Kyrpides NC, Tringe SG, Cavicchioli R (2013) High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc Natl Acad Sci USA 110:16939–16944
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485
Franzmann PD, Stackebrandt E, Sanderson K, Volkman JK, Cameron DE, Stevenson PL, Mcmeekin TA, Burton HR (1988) Halobacterium lacusprofundi sp. nov., a halophilic bacterium isolated from Deep Lake, Antarctica. Syst Appl Microbiol 11:20–27
González C, Gutiérrez C, Ramírez C (1978) Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Gupta RS, Naushad S, Baker S (2015) Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int J Syst Evol Microbiol 65:1050–1069
Gutiérrez C, González C (1972) Method for simultaneous detection of proteinase and esterase activities in extremely halophilic bacteria. Appl Microbiol 24:516–517
Gutiérrez MC, Castillo AM, Pagaling E, Heaphy S, Kamekura M, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A (2008) Halorubrum kocurii sp. nov., an archaeon isolated from a saline lake. Int J Syst Evol Microbiol 58:2031–2035
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hartmann R, Sickinger HD, Oesterhelt D (1980) Anaerobic growth of halobacteria. Proc Natl Acad Sci USA 77:3821–3825
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol 3:208–218
McGenity T, Grant WD (1995) Transfer of Halobacterium saccharovorum, Halobacterium sodomense, Halobacterium trapanicum NRC 34021 and Halobacterium lacusprofundi to the genus Halorubrum gen. nov., as Halorubrum saccharovorum comb. nov., Halorubrum sodomense comb. nov., Halorubrum trapanicum comb. nov., and Halorubrum lacusprofundi comb. nov. Syst Appl Microbiol 18:237–243
McGenity TJ, Grant WD (2001) Genus VII. Halorubrum. Bergey’s manual of systematic bacteriology. Springer, New York, pp 320–324
McGenity TJ, Oren A (2012) Life in saline environments. In: Bell E (ed) Life at extremes: environments, organisms, and strategies for survival. CABI International, Wallingford, pp 402–437
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T (2010) Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B’ (rpoB’) gene. Int J Syst Evol Microbiol 60:2398–2408
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Oren A (2012) Taxonomy of the family Halobacteriaceae: a paradigm for changing concepts in prokaryote systematics. Int J Syst Evol Microbiol 62:263–271
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47:233–238
Owen RJ, Hill LR (1979) The estimation of base compositions, base pairing and genome size of bacterial deoxyribonucleic acids. In: Gibbs BM (ed) Identification methods for microbiologists. Academic Press, Cambridge, pp 217–296
Papke RT, Corral P, Ram-Mohan N, de la Haba RR, Sánchez-Porro C, Makkay A, Ventosa A (2015) Horizontal gene transfer, dispersal and haloarchaeal speciation. Life (Basel) 5:1405–1426
Papke RT, Koenig JE, Rodríguez-Valera F, Doolittle WF (2004) Frequent recombination in a saltern population of Halorubrum. Science 306:1928–1929
Smibert RM, Krieg NR (1981) General characterization. In: Murray RGE et al (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC, pp 409–443
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
We thank Prof. Dr. Zhu L. Yang from the Kunming Institute of Botany, Chinese Academy of Sciences, for the help in sample collection, and Prof. Dr. Hua Xiang and Mr. Hong-Can Liu from the Institute of Microbiology, Chinese Academy of Sciences, for good suggestions and doing the DNA-DNA hybridization. This work was supported by Grants from the National Natural Science Foundation of China (31460003), the Anhui Provincial Key Lab of the Conservation, and Exploitation of Biological Resources (591601), the Anhui Provincial Natural Science Research Project (KJ2017A318), and the Education Department of Anhui province (gxyqZD2017011).
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession numbers of the 16S rRNA gene, rpoB′, and ef-2 sequences of strain Y69T are KX376706, KX595317, and KX595312, respectively. Three supplementary figures for cell morphology, thin-layer chromatogram of polar lipids, and Neighbor-Joining phylogenetic tree of 16S rRNA gene, rpoB′, and ef-2 genes are available with the online supplementary materials.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, S., He, J., Zhang, J. et al. Halorubrum salsamenti sp. nov., a Novel Halophilic Archaeon Isolated from a Brine of Salt Mine. Curr Microbiol 74, 1358–1364 (2017). https://doi.org/10.1007/s00284-017-1325-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1325-8