Abstract
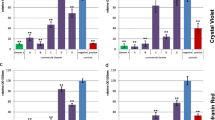
Biofilm protects bacteria from stress and hostile environment. Crystal violet (CV) assay is the most popular method for biofilm determination adopted by different laboratories so far. However, biofilm layer formed at the liquid–air interphase known as pellicle is extremely sensitive to its washing and staining steps. Early phase biofilms are also prone to damage by the latter steps. In bacteria like mycobacteria, biofilm formation occurs largely at the liquid–air interphase which is susceptible to loss. In the proposed protocol, loss of such biofilm layer was prevented. In place of inverting and discarding the media which can lead to the loss of the aerobic biofilm layer in CV assay, media was removed from the formed biofilm with the help of a syringe and biofilm layer was allowed to dry. The staining and washing steps were avoided, and an organic solvent—tetrahydrofuran (THF) was deployed to dissolve the biofilm, and the absorbance was recorded at 595 nm. The protocol was tested for biofilm estimation of E. coli, B. subtilis and M. smegmatis, and compared with the traditional CV assays. Isoniazid drug molecule, a known inhibitor of M. smegmatis biofilm, was tested and its inhibitory effects were quantified by the proposed protocol. For ease in referring, this method has been described as the Syal method for biofilm quantification. This new method was found to be useful for the estimation of early phase biofilm and aerobic biofilm layer formed at the liquid–air interphase. The biofilms formed by all three tested bacteria—B. subtilis, E. coli and M. smegmatis, were precisely quantified.



Similar content being viewed by others
References
Dufour D, Leung V, Lévesque CM (2010) Bacterial biofilm: structure, function, and antimicrobial resistance. Endod Top 22(1):2–16. doi:10.1111/j.1601-1546.2012.00277.x
Ferenci T (2016) Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol 24(3):209–223. doi:10.1016/j.tim.2015.11.009
Gonzalez Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK (2006) Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J Bacteriol 188(1):305–316. doi:10.1128/JB.188.1.305-316.2006
Hamon MA, Lazazzera BA (2001) The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42(5):1199–1209
Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR (2015) Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39(5):649–669
Mathew R, Mukherjee R, Balachandar R, Chatterji D (2006) Deletion of the rpoZ gene, encoding the ω subunit of RNA polymerase, results in pleiotropic surface-related phenotypes in Mycobacterium smegmatis. Microbiology 152(6):1741–1750. doi:10.1099/mic.0.28879-0
Naresh K, Avaji PG, Maiti K, Bharati BK, Syal K, Chatterji D, Jayaraman N (2012) Synthesis of beta-arabinofuranoside glycolipids, studies of their binding to surfactant protein-A and effect on sliding motilities of M. smegmatis. Glycoconj J 29(2–3):107–118. doi:10.1007/s10719-012-9369-2
O’Toole GA (2011) Microtiter dish biofilm formation assay. J Vis Exp 47:2437
O’Toole GA, Kolter R (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28(3):449–461
Schulze-Robbecke R, Janning B, Fischeder R (1992) Occurrence of mycobacteria in biofilm samples. Tuber Lung Dis 73(3):141–144
Steenackers HP, Parijs I, Foster KR, Vanderleyden J (2016) Experimental evolution in biofilm populations. FEMS Microbiol Rev 40(3):373–397
Syal K, Bhardwaj N, Chatterji D (2017) Vitamin C targets (p)ppGpp synthesis leading to stalling of long-term survival and biofilm formation in Mycobacterium smegmatis. FEMS Microbiol Lett. doi:10.1093/femsle/fnw282
Syal K, Chatterji D (2015) Differential binding of ppGpp and pppGpp to E. coli RNA polymerase: photo-labeling and mass spectral studies. Genes Cells 20(12):1006–1016. doi:10.1111/gtc.12304
Syal K, Flentie K, Bhardwaj N, Maiti K, Jayaraman N, Stallings CL, Chatterji D (2017) Synthetic (p)ppGpp analogue is an inhibitor of stringent response in mycobacteria. Antimicrob Agents Chemother. doi:10.1128/AAC.00443-17
Syal K, Joshi H, Chatterji D, Jain V (2015) Novel pppGpp binding site at the C-terminal region of the Rel enzyme from Mycobacterium smegmatis. FEBS J 282(19):3773–3785. doi:10.1111/febs.13373
Syal K, Maiti K, Naresh K, Avaji PG, Chatterji D, Jayaraman N (2016) Synthetic arabinomannan glycolipids impede mycobacterial growth, sliding motility and biofilm structure. Glycoconj J. doi:10.1007/s10719-016-9670-6
Syal K, Maiti K, Naresh K, Chatterji D, Jayaraman N (2015) Synthetic glycolipids and (p)ppGpp analogs: development of inhibitors for mycobacterial growth, biofilm and stringent response. Adv Exp Med Biol 842:309–327. doi:10.1007/978-3-319-11280-0_20
Teng R, Dick T (2003) Isoniazid resistance of exponentially growing Mycobacterium smegmatis biofilm culture. FEMS Microbiol Lett 227(2):171–174. doi:10.1016/s0378-1097(03)00584-6
Acknowledgement
KS was supported by the BASP Centre, Department of Biology, University of Copenhagen, Denmark and Indian Institute of Science, Bangalore, India, and is a recipient of Ranbaxy Science Scholar award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest to declare.
Rights and permissions
About this article
Cite this article
Syal, K. Novel Method for Quantitative Estimation of Biofilms. Curr Microbiol 74, 1194–1199 (2017). https://doi.org/10.1007/s00284-017-1304-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1304-0