Abstract
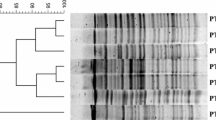
Although much is known about the mechanisms affecting cholera spread, cholera outbreaks occur annually in Iran. The aim of this study was to characterize and assess the clonal correlation of strains obtained from an outbreak in 2013 in Iran. Thirty-three strains of Vibrio cholerae were isolated from stool sample of patients majority of them belonged to Afghan nationality. PCR and sequencing analysis was performed to characterize virulence and resistance associates genes and cassettes. Clonality of isolates was assessed by Pulsed-field gel electrophoresis (PFGE) method. The ctx, zot, and tcp genes were present in 100 % of isolates. The wbeT gene was absent in all V. cholerae outbreak isolates, integrity of which is essential for Ogawa phenotype. This correlates with Inaba phenotype of all isolates under study. Sequencing of the ctxB + strains revealed that all isolates (El Tor strains) possessed the ctxB sequence of classical biotype allele known as El Tor variant strains. No class 1 or 2 integrons were detected among the isolates which indicate that in spite of high rate of resistance, integrons do not play an important role in V. cholerae resistance. All isolates were chloramphenicol sensitive all of which showed resistance to tetracycline and harbored the tetB resistance gene. PFGE analysis showed identical pulsotypes indicative of clonal dissemination of a single V. cholerae strain among the patients under study. Clonal cholera outbreak in boarder cities is alarming due to fear of import and spread of V. cholerae strains from out of the country which may lead to more spreading epidemics.

Similar content being viewed by others
References
Adabi M, Bakhshi B, Goudarzi H, Zahraei SM, Pourshafie MR (2009) Distribution of class I integron and sulfamethoxazole trimethoprim constin in Vibrio cholerae isolated from patients in Iran. Microb Drug Resist 15(3):179–184. doi:10.1089/mdr.2009.0885
Albert MJ, Rotimi VO, Dhar R, Silpikurian S, Pacsa AS, Molla AM, Szucs G (2009) Diarrhoeagenic Escherichia coli are not a significant cause of diarrhoea in hospitalised children in Kuwait. BMC Microbiol 9:62. doi:10.1186/1471-2180-9-62
Aliabad NH, Bakhshi B, Pourshafie MR, Sharifnia A, Ghorbani M (2012) Molecular diversity of CTX prophage in Vibrio cholerae. Lett Appl Microbiol 55(1):27–32. doi:10.1111/j.1472-765X.2012.03253.x
Ansaruzzaman M, Bhuiyan N, Nair GB, Sack DA, Lucas M, Deen JL, Ampuero J, Chaignat C-L, Group MCVDPC (2004) Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis 10(11):2057
Bakhshi B, Barzelighi HM, Adabi M, Lari AR, Pourshafie MR (2009) A molecular survey on virulence associated genotypes of non-O1 non-O139 Vibrio cholerae in aquatic environment of Tehran, Iran. Water Res 43(5):1441–1447. doi:10.1016/j.watres.2008.12.025
Bakhshi B, Boustanshenas M, Mahmoudi-Aznaveh A (2014) Emergence of Vibrio cholerae O1 classical biotype in 2012 in Iran. Lett Appl Microbiol 58(2):145–149. doi:10.1111/lam.12167
Bakhshi B, Pourshafie MR (2009) Assessing clonality of Vibrio cholerae strains isolated during four consecutive years (2004–2007) in Iran. Scand J Infect Dis 41(4):256–262. doi:10.1080/00365540902767049
Bakhshi B, Pourshafie MR, Navabakbar F, Tavakoli A (2008) Genomic organisation of the CTX element among toxigenic Vibrio cholerae isolates. Clin Microbiol Infect 14(6):562–568. doi:10.1111/j.1469-0691.2008.01976.x
Basu A, Garg P, Datta S, Chakraborty S, Bhattacharya T, Khan A, Ramamurthy S, Bhattacharya SK, Yamasaki S, Takeda Y, Nair GB (2000) Vibrio cholerae O139 in Calcutta, 1992-1998: incidence, antibiograms, and genotypes. Emerg Infect Dis 6(2):139–147. doi:10.3201/eid0602.000206
Cambray G, Guerout AM, Mazel D (2010) Integrons. Annu Rev Genet 44:141–166. doi:10.1146/annurev-genet-102209-163504
Chakraborty S, Garg P, Ramamurthy T, Thungapathra M, Gautam JK, Kumar C, Maiti S, Yamasaki S, Shimada T, Takeda Y, Ghosh A, Nair GB (2001) Comparison of antibiogram, virulence genes, ribotypes and DNA fingerprints of Vibrio cholerae of matching serogroups isolated from hospitalised diarrhoea cases and from the environment during 1997-1998 in Calcutta, India. J Med Microbiol 50(10):879–888
Choopun N, Louis V, Huq A, Colwell RR (2002) Simple procedure for rapid identification of Vibrio cholerae from the aquatic environment. Appl Environ Microbiol 68(2):995–998
Chun J, Huq A, Colwell RR (1999) Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl Environ Microbiol 65(5):2202–2208
Cooper K, Luey C, Bird M, Terajima J, Nair G, Kam K, Arakawa E, Safa A, Cheung D, Law C (2006) Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog Dis 3(1):51–58
Dalsgaard A, Forslund A, Bodhidatta L, Serichantalergs O, Pitarangsi C, Pang L, Shimada T, Echeverria P (1999) A high proportion of Vibrio cholerae strains isolated from children with diarrhoea in Bangkok, Thailand are multiple antibiotic resistant and belong to heterogenous non-O1, non-O139 O-serotypes. Epidemiol Infect 122(2):217–226
Dalsgaard A, Forslund A, Serichantalergs O, Sandvang D (2000) Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob Agents Chemother 44(5):1315–1321
Das S, Saha R, Kaur IR (2008) Trend of antibiotic resistance of Vibrio cholerae strains from East Delhi. Indian J Med Res 127(5):478–482
Dashtbani-Roozbehani A, Bakhshi B, Pourshafie MR (2013) Genetic relatedness of clinical and environmental Vibrio cholerae isolates based on triple housekeeping gene analysis. Curr Microbiol 67(1):15–20. doi:10.1007/s00284-013-0324-7
Furushita M, Shiba T, Maeda T, Yahata M, Kaneoka A, Takahashi Y, Torii K, Hasegawa T, Ohta M (2003) Similarity of tetracycline resistance genes isolated from fish farm bacteria to those from clinical isolates. Appl Environ Microbiol 69(9):5336–5342
Hajia M, Rahbar M, Rahnamye Farzami M, Masoumi Asl H, Dolatyar A, Imani M, Saburian R, Mafi M, Bakhshi B (2015) Assessing Clonal Correlation of Epidemic Vibrio cholerae Isolates During 2011 in 16 Provinces of Iran. Curr Microbiol 70(3):408–414. doi:10.1007/s00284-014-0725-2
Hall RM, Collis CM (1995) Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol 15(4):593–600
Iyer A, Barbour E, Azhar E, Salabi AAE, Hani Mutlak A, Hassan IQ, Adeel Chaudhary AA, Kumosani T, Damanhouri G, Alawi M, Na’was T, Nour AMA, Harakeh S (2013) Transposable elements in Escherichia coli antimicrobial resistance. Adv Biosci Biotechnol 4:415–423
Jain M, Goel AK, Bhattacharya P, Ghatole M, Kamboj DV (2011) Multidrug resistant Vibrio cholerae O1 El Tor carrying classical ctxB allele involved in a cholera outbreak in South Western India. Acta Trop 117(2):152–156. doi:10.1016/j.actatropica.2010.12.002
Kaper JB, Morris JG Jr, Levine MM (1995) Cholera. Clin Microbiol Rev 8(1):48–86
Karaolis DK, Lan R, Reeves PR (1995) The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol 177(11):3191–3198
Kim YH, Jun LJ, Park SH, Yoon SH, Chung JK, Kim JC, Jeong HD (2007) Prevalence of tet(B) and tet(M) genes among tetracycline-resistant Vibrio spp. in the aquatic environments of Korea. Dis Aquat Organ 75(3):209–216. doi:10.3354/dao075209
Kumar P, Jain M, Goel A, Bhadauria S, Sharma S, Kamboj D, Singh L, Ramamurthy T, Nair G (2009) A large cholera outbreak due to a new cholera toxin variant of the Vibrio cholerae O1 El Tor biotype in Orissa. Eastern India. J Med Microbiol 58(2):234–238
Mazel D, Davies J (1999) Antibiotic resistance in microbes. Cell Mol Life Sci 56(9–10):742–754
Mercy N, Mohamed AA, Zipporah N, Chowdhury G, Pazhani GP, Ramamurthy T, Boga HI, Kariuki SM, Joseph O (2014) Phenotypic and genetic characterization of vibrio cholerae O1 isolated from various regions of Kenya between 2007 and 2010. Pan Afr Med J 19:8. doi:10.11604/pamj.2014.19.8.2496
Morita M, Ohnishi M, Arakawa E, Yamamoto S, Nair GB, Matsushita S, Yokoyama K, Kai A, Seto K, Watanabe H, Izumiya H (2010) Emergence and genetic diversity of El Tor Vibrio cholerae O1 that possess classical biotype ctxB among travel-associated cases of cholera in Japan. J Med Microbiol 59(Pt 6):708–712. doi:10.1099/jmm.0.017624-0
Mukhopadhyay AK, Chakraborty S, Takeda Y, Nair GB, Berg DE (2001) Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae. J Bacteriol 183(16):4737–4746. doi:10.1128/jb.183.16.4737-4746.2001
Nair GB, Qadri F, Holmgren J, Svennerholm AM, Safa A, Bhuiyan NA, Ahmad QS, Faruque SM, Faruque AS, Takeda Y, Sack DA (2006) Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol 44(11):4211–4213. doi:10.1128/JCM.01304-06
Ng LK, Mulvey MR, Martin I, Peters GA, Johnson W (1999) Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob Agents Chemother 43(12):3018–3021
Nguyen BM, Lee JH, Cuong NT, Choi SY, Hien NT, Anh DD, Lee HR, Ansaruzzaman M, Endtz HP, Chun J, Lopez AL, Czerkinsky C, Clemens JD, Kim DW (2009) Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol 47(5):1568–1571. doi:10.1128/JCM.02040-08
Olsvik O, Wahlberg J, Petterson B, Uhlen M, Popovic T, Wachsmuth IK, Fields PI (1993) Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J Clin Microbiol 31(1):22–25
Perilla M, Ajello G, Bopp C, Elliott J, Facklam R (2003) Manual for the laboratory identification and antimicrobial susceptibility testing of bacterial pathogens of public health importance in the developing world. Haemophilus influenzae Neisseria meningitidis Streptococcus pneumoniae Neisseria gonorrhoeae Salmonella serotype Typhi Shigella and Vibrio cholerae
Popovic T, Fields PI, Olsvik O (1994) Detection of cholera toxin genes. Vibrio cholerae and cholera: molecular to global perspectives. American Society for Microbiology, Washington, pp 41–52
Popovic TFP, Olsvik O (1994) Vibrio cholerae and cholera: molecular to global perspectives. In: Wachsmuth IK, Blake PA, Olsvik O (eds) Vibrio cholerae and cholera: molecular to global perspectives. American Society for Microbiology, Washington, pp 41–52
Pourshafie M, Grimont F, Kohestani S, Grimont PA (2002) A molecular and phenotypic study of Vibrio cholerae in Iran. J Med Microbiol 51(5):392–398
Pourshafie MR, Bakhshi B, Ranjbar R, Sedaghat M, Sadeghifard N, Zaemi Yazdi J, Parzadeh M, Raesi J (2007) Dissemination of a single Vibrio cholerae clone in cholera outbreaks during 2005 in Iran. J Med Microbiol 56(Pt 12):1615–1619. doi:10.1099/jmm.0.47218-0
Pourshafie MR, Grimont F, Saifi M, Grimont PA (2000) Molecular epidemiological study of Vibrio cholerae isolates from infected patients in Teheran, Iran. J Med Microbiol 49(12):1085–1090
Rahmani F, Fooladi AAI, Marashi SMA, Nourani MR (2012) Drug resistance in Vibrio cholerae strains isolated from clinical specimens. Acta Microbiol Immunol Hung 59(1):77–84
Ramamurthy T, Yamasaki S, Takeda Y, Nair GB (2003) Vibrio cholerae O139 Bengal: odyssey of a fortuitous variant. Microbes Infect 5(4):329–344
Ranjbar M, Rahmani E, Nooriamiri A, Gholami H, Golmohamadi A, Barati H, Rajabifar D, Barati S, Sabet MS, Zamiri A, Haghighi S, Taifehashemi P, Nojomi M (2010) High prevalence of multidrug-resistant strains of Vibrio cholerae, in a cholera outbreak in Tehran-Iran, during June-September 2008. Trop Doct 40(4):214–216. doi:10.1258/td.2010.100015
Raychoudhuri A, Patra T, Ghosh K, Ramamurthy T, Nandy RK, Takeda Y, Balakrish-Nair G, Mukhopadhyay AK (2009) Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg Infect Dis 15(1):131–132. doi:10.3201/eid1501.080543
Safa A, Sultana J, Dac Cam P, Mwansa JC, Kong RY (2008) Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg Infect Dis 14(6):987–988. doi:10.3201/eid1406.080129
Sedaghat M, Rahimi F, Talebi M, Pourshafie MR (2013) Serotyping, Antibiotic Susceptibility Pattern and Detection of hlyA Gene Among Cholera Patients in Iran. Jundishapur J Microbiol 6(1):20–23
Sharifnia A, Bakhshi B, Pourshafie MR (2012) wbeT sequence typing and IS1004 profiling of Vibrio cholerae isolates. Lett Appl Microbiol 54(4):267–271. doi:10.1111/j.1472-765X.2012.03204.x
Shi L, Fujihara K, Sato T, Ito H, Garg P, Chakrabarty R, Ramamurthy T, Nair GB, Takeda Y, Yamasaki S (2006) Distribution and characterization of integrons in various serogroups of Vibrio cholerae strains isolated from diarrhoeal patients between 1992 and 2000 in Kolkata, India. J Med Microbiol 55(Pt 5):575–583. doi:10.1099/jmm.0.46339-0
Stavric S, Bachanan B (1995) The Isolation and Identification of V. cholerae O1 and non-O1 from Foods. Polyscience Publication, Government of Canada, Health Protection Branch, Ottawa (MFLP-72), Canada
Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, Joyce K, Turnsek M, Garrett N, Humphrys M (2011) Characterization of toxigenic Vibrio cholerae from Haiti, 2010–2011. Emerg Infect Dis 17(11):2122
Tamayo M, Koblavi S, Grimont F, Castaneda E, Grimont P (1997) Molecular epidemiology of Vibrio cholerae O1 isolates from Colombia. J Med Microbiol 46(7):611–616
Waters SH, Rogowsky P, Grinsted J, Altenbuchner J, Schmitt R (1983) The tetracycline resistance determinants of RP1 and Tn1721: nucleotide sequence analysis. Nucleic Acids Res 11(17):6089–6105
White PA, McIver CJ, Rawlinson WD (2001) Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother 45(9):2658–2661
Yamamoto T, Nair GB, Takeda Y (1995) Emergence of tetracycline resistance due to a multiple drug resistance plasmid in Vibrio cholerae O139. FEMS Immunol Med Microbiol 11(2):131–136
Yu L, Zhou Y, Wang R, Lou J, Zhang L, Li J, Bi Z, Kan B (2012) Multiple antibiotic resistance of Vibrio cholerae serogroup O139 in China from 1993 to 2009. PLoS One 7(6):e38633
Acknowledgments
This study was supported by a Grant from Research council of Tarbiat Modares University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bakhshi, B., Mahmoudi-Aznaveh, A. & Salimi-Khorashad, A. Clonal Dissemination of a Single Vibrio cholerae O1 Biotype El Tor Strain in Sistan–Baluchestan Province of Iran During 2013. Curr Microbiol 71, 163–169 (2015). https://doi.org/10.1007/s00284-015-0806-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0806-x