Abstract
A novel Gram-positive, anaerobic, and coccus-shaped bacterium, designated as strain ChDC B134T, was isolated from a human postoperative infectious lesion in the right maxilla between the lateral incisor and canine and was characterized by polyphasic taxonomic analysis. 16S rRNA gene sequence analysis revealed that the strain ChDC B134T belonged to the genus Peptoniphilus, as it showed sequence similarities to Peptoniphilus indolicus KCTC 15023T (94.0 %) and Peptoniphilus asaccharolyticus KCTC 3321T (93.8 %). The prevalent fatty acids of of strain ChDC B134T were C16:0 (20.3 %), C18:1 cis 9 (34.3 %), and C18:0 (13.2 %). The DNA G+C content was 30.9 mol%. The cell wall diamino acid was d-ornithine, which is a property shared by other reference type strains of the genus Peptoniphilus. Based on the results of phenotypic, chemotaxonomic, and phylogenetic analysis, strain ChDC B134T (=KCOM 1628T = KCTC 15227T = JCM 30223T) should be classified as the type strain of a novel species of genus Peptoniphilus, for which the name Peptoniphilus mikwangii sp. nov. is proposed.


Similar content being viewed by others
References
Byström A, Sundqvist G (1981) Bacteriologic evaluation of the efficacy of mechanical root canal instrumentation in endodontic therapy. Scand J Dent Res 89:321–328
Citron DM, Tyrrell KL, Goldstein EJ (2012) Peptoniphilus coxii sp. nov. and Peptoniphilus tyrrelliae sp. nov. isolated from human clinical infections. Anaerobe 18:244–248. doi:10.1016/j.anaerobe.2011.11.008
Conrads G, Soffner J, Pelz K, Mutters R (1997) Taxonomic update and clinical significance of species within the genus Peptostreptococcus. Clin Infect Dis 25(Suppl 2):S94–S97
Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S (2001) Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol 51:1521–1528
Ezaki T, Yamamoto N, Ninomiya K, Suzuki S, Yabuuchi E (1983) Transfer of Peptococcus indolicus, Peptococcus asaccharolyticus, Peptococcus prevotii, and Peptococcus magnus to the Genus Peptostreptococcus and proposal of Peptostreptococcus tetradius sp. nov. Int J Syst Evol Microbiol 33:683–698
Gajan EB, Aghazadeh M, Abashov R, Salem Milani A, Moosavi Z (2009) Microbial flora of root canals of pulpally-infected teeth: Enterococcus faecalis a prevalent species. J Dent Res Dent Clin Dent Prospects 3:24–27. doi:10.5681/joddd.2009.007
Gomes BP, Lilley JD, Drucker DB (1996) Clinical significance of dental root canal microflora. J Dent 24:47–55
Gomes BP, Pinheiro ET, Jacinto RC, Zaia AA, Ferraz CC, Souza-Filho FJ (2008) Microbial analysis of canals of root-filled teeth with periapical lesions using polymerase chain reaction. J Endod 34:537–540. doi:10.1016/j.joen.2008.01.016
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Li N, Hashimoto Y, Adnan S, Miura H, Yamamoto H, Ezaki T (1992) Three new species of the genus Peptostreptococcus isolated from humans: Peptostreptococcus vaginalis sp. nov., Peptostreptococcus lacrimalis sp. nov., and Peptostreptococcus lactolyticus sp. nov. Int J Syst Bacteriol 42:602–605
Mishra AK, Lagier JC, Nguyen TT, Raoult D, Fournier PE (2013) Non contiguous-finished genome sequence and description of Peptoniphilus senegalensis sp. nov. Stand Genomic Sci 7:370–381. doi:10.4056/sigs.3366764
Murdoch DA (1998) Gram-positive anaerobic cocci. Clin Microbiol Rev 11:81–120
Murdoch DA, Shah HN (1999) Reclassification of Peptostreptococcus magnus (Prevot 1933) Holdeman and Moore 1972 as Finegoldia magna comb. nov. and Peptostreptococcus micros (Prevot 1933) Smith 1957 as Micromonas micros comb. nov. Anaerobe 5:555–559
Rooney AP, Swezey JL, Pukall R, Schumann P, Spring S (2011) Peptoniphilus methioninivorax sp. nov., a Gram-positive anaerobic coccus isolated from retail ground beef. Int J Syst Evol Microbiol 61:1962–1967. doi:10.1099/ijs.0.024232-0
Schleifer KH (1985) Analysis of the chemical composition and primary structure of murein. Methods Microbiol 18:123–156
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849. doi:10.1099/00207713-44-4-846
Song Y, Liu C, Finegold SM (2007) Peptoniphilus gorbachii sp. nov., Peptoniphilus olsenii sp. nov., and Anaerococcus murdochii sp. nov. isolated from clinical specimens of human origin. J Clin Microbiol 45:1746–1752
Sundqvist G (1992) Associations between microbial species in dental root canal infections. Oral Microbiol Immunol 7:257–262
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tindall BJ, Euzéby JP (2006) Proposal of Parvimonas gen. nov. and Quatrionicoccus gen. nov. as replacements for the illegitimate, prokaryotic, generic names Micromonas Murdoch and Shah 2000 and Quadricoccus Maszenan et al. 2002, respectively. Int J Syst Evol Microbiol 56:2711–2713
Ulger-Toprak N, Lawson PA, Summanen P, O’Neal L, Finegold SM (2012) Peptoniphilus duerdenii sp. nov. and Peptoniphilus koenoeneniae sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol 62:2336–2341. doi:10.1099/ijs.0.031997-0
Williams BL, McCann GF, Schoenknecht FD (1983) Bacteriology of dental abscesses of endodontic origin. J Clin Microbiol 18:770–774
Yoo SY, Kim KJ, Lim SH, Kim KW, Hwang HK, Min BM, Choe SJ, Kook JK (2005) First isolation of Streptococcus downei from human dental plaques. FEMS Microbiol Lett 249:323–326
Acknowledgments
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2013M3A9B8013860) and in part by research funds from Chosun University, 2013.
Conflict of interest
The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Eugene Cho and Soon-Nang Park have contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
284_2014_712_MOESM1_ESM.pptx
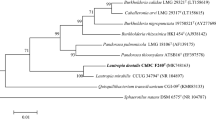
Supplementary Fig. S1. Maximum-likelihood (a) and maximum-parsimony (b) phylogenetic trees based on 16S rDNA sequences showing the relationship between strain ChDC B134T and related species. The stability of the phylogenetic trees was assessed by bootstrap analysis of 1,000 replicates using MEGA version 6.06 [25]. Bar, 0.01 changes per nucleotide position. (PPTX 110 kb)
284_2014_712_MOESM2_ESM.pptx
Supplementary Fig. S2. Thin-layer chromatography of acid hydrolysates of peptidoglycan. Strains: 1, strain ChDC B134T; 2, P. asaccharolyticus KCTC 3321T; 3, P. indolicus KCTC 15023T; 4, P. lacrimalis KCTC 5950T. d-orn, d-ornithine (red-colored arrow); l-orn, l-ornithine (green-colored arrow). (PPTX 298 kb)
Rights and permissions
About this article
Cite this article
Cho, E., Park, SN., Shin, Y. et al. Peptoniphilus mikwangii sp. nov., Isolated from a Clinical Specimen of Human Origin. Curr Microbiol 70, 260–266 (2015). https://doi.org/10.1007/s00284-014-0712-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0712-7