Abstract
Partial 18S rRNA gene sequences of the three trichodinids, namely Trichodina modesta Lom, 1970, Trichodina paraheterodentata Tang and Zhao 2012. and Trichodinella epizootica (Raabe 1950) Šrámek-Hušek, 1953, were acquired and used to construct phylogenetic trees. The results revealed that Trichodinella epizootica clustered with Trichodinella sp.; Trichodina paraheterodentata Tang and Zhao 2012 was sister to the clade composed of Trichodina heterodentata Duncan, 1977 and Trichodina nobilis Chen, 1963; Trichodina modesta Lom, 1970 clustered with Trichodina reticulata Hirschman and Partsch, 1955. The branching order of species within the Mobilia clade was closely correlated with GC content. Furthermore, blade morphology was also found to be the primary morphological character in determining the phylogenetic relationships among members of the genus Trichodina. The present findings suggest that the genus Trichodina is paraphyletic when species of Trichodinella are included in the analyses.
Similar content being viewed by others
Introduction
Members of the family Trichodinidae are best known as ectoparasites of fishes. About 300 species of trichodinids have been described from fishes, mostly from freshwater environments [35]. In China, the trichodinid ciliates of freshwater fishes have received considerable attention in recent years [12, 13, 22–31, 36, 39–42]. Hitherto, most studies have focused on their morphology following silver impregnation. However, morphological characters have proved inadequate to reconstruct evolutionary history as many are unique to the sub-class Mobilia so their weighting is difficult to determine and some, such as the presence or absence of central granules in the adhesive disc, lack a consensus as to their systematic importance. Molecular data are increasingly used for studying phylogenetic relationships among ciliates. However, there have been relatively few such studies of mobilians prompting calls for more sequence data for taxa within this group [8, 9, 33, 37].
In this article, we sequenced the small subunit rRNA (18S rRNA) gene of three trichodinids, namely Trichodina modesta Lom, 1970; Trichodina paraheterodentata Tang and Zhao, 2012 and Trichodinella epizootica (Raabe 1950) Šrámek-Hušek 1953, in order to analyse their molecular phylogeny. The main aims of this work are to increase knowledge and understanding of the diversity and phylogeny of trichodinids. The importance of denticle blade morphology, central granules and GC content in the phylogeny of trichodinids are also discussed.
Materials and Methods
Collection and Identification (Fig. 1)
Specimens of host fishes, Siniperca chuatsi (ca. 1 year old, 15–35 cm in length), Misgurnus anguillicaudatus (ca. 1 year old, 10–26 cm in length) and Carassius auratus (ca. 1 year old, 8–25 cm in length) were collected from the Jialing River in the urban zone of Chongqing, China between February 2005 and April 2009. Each host was necropsied and examined under a binocular dissecting microscope (NIKON SMZ1500) at 400× in order to detect trichodinids. Fresh gill or skin smears containing trichodinids were prepared and impregnated using the dry silver method of Klein [11]. The nuclear apparatus was revealed using the methyl green-pyronin stain [6]. Observations, counts and measurements on impregnated specimens were performed using a compound microscope (NIKON E600, Nikon Instrument Inc., Shanghai, China) at a magnification of 1,000 × . Systematics follows Lynn (2008) [17] and Zhan et al. (2009) [37]. Terminology is mainly according to Corliss (1979) [4].
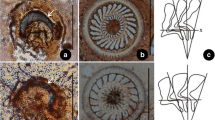
Photomicrographs of silver-impregnated adhesive discs of three trichodinids. A–B Trichodina paraheterodentata Tang and Zhao 2012 (from Siniperca chuatsi); C Trichodina modesta Lom, 1970 (from Misgurnus anguillicaudatus); D Trichodinella epizootica (Raabe,1950) Šrámek-Hušek 1953 (from Carassius auratus). (Scale bar 20 μm)
DNA Extraction, Amplification, Cloning, and Sequencing
For each trichodinid species, at least 4 or 5 individuals were harvested, washed several times in a PCR tube and centrifuged at 6000–7500×g. DNA was extracted using REDExtract-N-AmpTM Tissue PCR Kit (Sigma, St. Louis, USA) following the manufacturer’s instructions.
The 18S rRNA genes of T. paraheterodentata and T. epizootica were amplified by the polymerase chain reaction (PCR) with the universal eukaryotic primers, forward primer 5′-AAC CTG GTT GAT CCT GCC AGT-3′, reverse primer 5′-TGA TCC TTC TGC AGG TTC ACC TAC-3′ [18]. Temperature cycling was five cycles of denaturation for 1 min at 94 °C, primer annealing for 2 min at 56 °C, and extension for 2 min at 72 °C, followed by 35 cycles in the same manner, but with the annealing temperature increased to 62 °C, and a final extended elongation step at 72 °C for 10 min. The 18S rRNA gene of T. modesta, was amplified with the primer pair MX5-MX3, forward primer 5′-CTG CGG ACG GCT CAGTAA ATC AGT-3′ and reverse primer 5′-CCA GGA CAT CTT AGG GCA TCA CAGA-3′ [1]. The cycling parameters were as follows: 5 min initial denaturation at 94 °C; then 35 cycles of 1 min at 94 °C, 1 min at 56 °C, and 2 min at 72 °C, followed by an extended elongation step at 72 °C for 10 min. Purified PCR products were inserted into a pMD18-T vector (TaKaRa) and selected clones were sequenced in an ABI Prism 377 DNA Sequencer (Applied Biosystems Inc., Foster City, California).
Phylogenetic Analyses
The nucleotide sequences used for the present analyses are available from GenBank databases (for accession numbers see Table 1). A total of 24 complete or partial 18S rRNA gene sequences, including those of our three newly sequenced trichodinid species, were used to construct the phylogenetic trees. The hypotrich Euplotes minuta was the outgroup taxon. All sequences were first aligned using Clustal X 1.81 [32] and further modified manually using BioEdit 5.0.6 [10] with consideration of the secondary structures. Maximum likelihood (ML) and Bayesian Inference analyses were employed for tree construction. The ML tree was constructed in PAUP*4.0b10 [21]. Bootstrap confidence values were calculated with a heuristic search using simple sequence addition and 100 replicates. Bayesian analyses were conducted in MrBayes 3.1.2 [19] under a GTR model with 106 generations, tree sampling every 100 generations, with a burn-in of 10000 trees to generate a posterior probability distribution using Markov chain Monte Carlo (MCMC) methods.
Results
GC content Analyses (Fig. 2, Table 2)
The GC contents of 18S rDNA for the 11 mobilian species used in the phylogenetic analyses are listed in Table 2. Five trichodinids have a GC content more than 50 %, namely T. paraheterodentata, T. heterodentata, T. nobilis, Trichodinella sp. and T. epizootica. Trichodinids with GC content between 48 and 50 % include the three marine species, Trichodina ruditapicis, T. sinonovaculae and T. meretricis. Two trichodinids have GC content between 46 and 48 %, i.e. T. modesta and T. reticulata. Urceolaria urechi has the lowest GC content, between 44 and 46 %.
In the 18S rRNA gene trees, the branching order of the different clades corresponded with the GC content of species within each clade. For example, trichodinids with a GC content more than 50 %, i.e. T. paraheterodentata, T. heterodentata, T. nobilis, Trichodinella sp. and T. epizootica, clustered together in the terminal clade. These were preceded by a clade comprising three species, i.e. T. ruditapicis, T. sinonovaculae and T. meretricis, all of which have a GC content between 48 and 50 %. The clade that branched first within the trichodinid clade comprises two species, viz. T. modesta and T. reticulata, both of which have a GC content between 46 and 48 %. U. urechi, which branched basally within the Mobilia, possesses the lowest GC content, between 44 and 46 %. (Fig. 3).
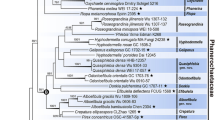
The phylogenetic tree of selected species of oligohymenophorean sub-classes Peritrichia s. str. and Mobilia inferred from small subunit rDNA sequences using maximum likelihood (ML) by Paup.4.10 and Bayesian analysis with the model of ‘‘GTR+1+G’’. The numbers at the nodes represent the bootstrap percentages from 1,000 replicates for ML and the posterior probabilities from 1,000,000 generations for Bayesian analysis, respectively
Phylogenetic Analyses (Fig. 3)
The phylogenetic trees, based on maximum likelihood and Bayesian inference analyses, respectively, had very similar topologies therefore only one tree is presented here (Fig. 3). This reveals that (1) the 23 species of Peritrichia s. str. and Mobilia were divided into two distinctly separate branches; (2) the urceolariid clade, represented by U. urechi, branched basally within the Mobilia and was sister to the trichodinid assemblage; (3) the two Trichodinella species clustered together in a clade nested within the trichodinid assemblage which otherwise comprises only species of Trichodina, suggesting the latter is paraphyletic; (4) the two species of Trichodina with central granules in the adhesive disc, namely T. modesta and T. reticulata, clustered together in a basal position within the trichodinid assemblage; (5) those species from marine mollusc hosts, i.e. Trichodina sinonovaculae, T. meretricis and T. ruditapicis, clustered together in a clade nested within the remaining trichodinids, all of which were isolated from freshwater fish hosts; (6) T. paraheterodentata did not cluster with T. heterodentata in any of the analyses but rather was sister to the clade comprising T. heterodentata and T. nobilis.
Discussion
In the 18S rRNA gene trees, the peritrichs s.l. were divided into two well-supported clades: the sessilid forms or Peritrichia s.str. and the mobilian forms, which are now recognised as the sub-class Mobilia [37]. This finding is consistent with previous phylogenetic analyses of peritrichs based on gene sequence data such as 18S rRNA and α-tubuline [8, 9, 33, 37]. Representatives of three genera of mobilians were included in the present analyses: Urceolaria, Trichodinella and Trichodina. Urceolaria branched separately from the rest of the Mobilia, which was expected since it belongs to the family Urceloariidae as opposed to all other taxa in the analyses which are members of the family Trichodinidae. By contrast, the Trichodinella clade nested within the Trichodina assemblage. Trichodinella is separated from Trichodina by two main morphological characters: the development of the adoral rows of cilia which turn 180°–270° around the peristome in the former (vs. 360°–540° in Trichodina) and the denticles which have short, stunted thorns and delicate blades (vs. denticles robust with well-developed thorns and blades in Trichodina). These differences have long been considered sufficient for generic separation [15, 16].
Hitherto, gene sequence datum was available for only one species of Trichodenella, viz. Trichodinella.sp., which was ever reported by the name of T. myakkae [9]. This species has been included in at least two previous studies of mobilian phylogeny based on 18S rRNA gene sequence and in both cases it was nested within a larger Trichodina assemblage [9, 33]. Consequently, this called into question the identity of the genus Trichodinella [33], and doubts about the identity of T. myakkae resulted in its exclusion from the analysis by Zhan et al. [37]. In this study, a second species of Trichodinella, viz. T. epizootica the identity of which was confirmed by careful morphological examination (Fig. 1), was sequenced for the first time. As expected, T. epizootica had a high level of similarity (97 %) with Trichodinella sp. and the two clustered together with maximum bootstrap support, suggesting that they are congeneric. The inclusion of a second species of Trichodinella made no difference to the placement of this genus in the gene tree.
One factor that has not previously been taken into account when considering phylogenetic relationships among mobilians is GC content. In this study, it was noted that the branching order of the various clades closely corresponded with the GC content of the constituent species, those with a lower GC content (e.g. U. urechi, GC content 44–46 %) branching first, with each successive clade having increasing GC content, those with the highest GC content (i.e. T. paraheterodentata Tang and Zhao 2012, T. heterodentata, T. nobilis, Trichodonella sp. and T. epizootica) branching last. The GC content is traditionally regarded as being characteristic of the genome of any given organism and, in the case of bacteria, has been used in taxonomy and classification [7, 20]. Furthermore, Du et al. [5] used the GC levels of genome-wide genes to determine the correlation between the GC content and evolutionary relationships. This is consistent with the findings of Zhang et al. [38] who also reported a close association between GC content and evolutionary relationships among lichens. The biological significance of GC content is not fully understood. For example, Cao et al. [3] unexpectedly discovered the function of lower GC content in editing exons and revealed a possible relationship between molecular characteristics of DNA, RNA and purifying selection. Clearly, the influence of GC content on trichodinid phylogeny needs further investigation. Thus, there remain four possible explanations for the placement of Trichodinella in the 18S rRNA gene tree (1) that Trichodinella and Trichodina should not be separated at the level of genus; (2) the genus Trichodina is paraphyletic; (3) the placement of Trichodinella is an artifact and its true phylogenetic position is not recovered in the present analysis due to using just a single gene, undersampling etc.; (4) the placement of Trichodinella reflects its GC content rather than its true phylogenetic position.
A morphological character that can be mapped onto the 18S rRNA gene tree with a high level of correlation is the shape of the denticle blade. Denticle morphology is an important character for the circumscription and identification of species and genera of trichodinids [2, 14, 34]. Therefore, it is not surprising that species with similar blade shapes tend to cluster together within the gene tree of the Mobilia (Figs. 4). For example, the three Trichodina species within the terminal clade, i.e. T. paraheterodentata, T. heterodentata and T. nobilis, all possess an arc-shaped blade; the two Trichodinella species, Trichodina sp. and T. epizootica, cluster together and both have long, strip-like blades; the three Trichodina species from marine mollusc hosts, T. meretricis, T. ruditapicis and T. sinovaculae, all have irregular quadrangular-shaped blades, and; the two species that branch basally within the trichodinid clade, T. modesta and T. reticulata, have a regular quadrangular-shaped blade (Fig. 4). These findings support the view that denticle blade shape is significant in the phylogeny of the Mobilia [37].
Denticle morphology of different trichodinids, arrows mark the blade of the denticle and the yellow lines indicate the Y-axis. A–B Trichodina reticulata (Tang and Zhao, 2010); C Trichodina modesta (present work); D Trichodina heterodentata (Gong et al. 2006); E Trichodina nobilis (Gong et al. 2006); F Trichodina paraheterodentata (Tang and Zhao 2012); G Trichodina sinonovaculae (Xu et al. 1999); H Trichodina meretricis (Xu et al. 1999); I Trichodina ruditapicis (Xu et al. 2000); J Trichodinella sp. (Gong et al. 2006); K Trichodinella epizootica (present work). (Scale bar 20 μm)
Another morphological character suggested as being of phylogenetic importance among mobilians is the presence or absence of central granules in the adhesive disc [9]. However, we could not find evidence to support this in this study, which is consistent with the findings of Zhan et al. [37]. For example T. reticulata, which possesses granules, is sister to T. modesta, which lacks granules. By contrast, there was evidence that phylogeny among mobilians may be influenced by the host and/or habitat with the three Trichodina species from marine mollusc hosts, T. meretricis, T. ruditapicis and T. sinovaculae, clustering together to the exclusion of the other species, all of which were isolated from freshwater fishes. This is consistent with Zhan et al. [37] who also remarked on the possible importance of co-evolution with the host in the phylogeny of mobilians.
Clearly gene sequence data are of growing importance in determining phylogenetic relationships among mobilians. However, undersampling remains a significant barrier to progress with sequence data being available for only 14 out of a possible ca. 300 mobilian species. Furthermore, with the notable exception of Gong et al. [8] who analysed the α-tubulin gene of 10 mobilian species, data are only available for the 18S rRNA gene. Thus, taxon sampling needs to be increased, and a wider range of genes analysed, before we can fully elucidate the phylogeny of the Mobilia.
References
Andree K, SzéKely C, Molná K, Gresoviac S, Hedrick R (1999) Relationships among members of the genus Myxobolus (Myxozoa: Bilvalvidae) based on small subunit ribosomal DNA sequences. J Parasitol 85:68–74
Arthur JR, Margolis L (1984) Trichodina truttae Müller, 1937 (Ciliophora: Peritrichida), a common pathogenic ectoparasite of cultured juvenile salmonid fishes in British Columbia: redescription and examination by scanning electron microscopy. Can J Zool 62:1842–1848
Cao J, Wu X, Jin Y (2008) Lower GC-content in editing exons: implications for regulation by molecular characteristics maintained by selection. Gene 421:14–19
Corliss JO (1979) The ciliated protozoa, 2nd edn. Pergamon, Oxford
Du H, Hu H, Meng Y, Zheng W, Ling F, Wang J, Zhang X, Nie Q, Wang X (2010) The correlation coefficient of GC content of the genome-wide genes is positively correlated with animal evolutionary relationships. FEBS Lett 584:3990–3994
Foissner W (1991) Basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa. Eur J Protistol 27:313–330
Garrity FL, Detrick B, Dennedy ER (1969) Deoxyribonucleic acid base composition in the taxonomy of Staphylococcus. J Bacteriol 97:557–560
Gong Y, Xu K, Zhan Z, Yu Y, Li X, Villalobo E, Feng W (2010) Alpha-tubulin and small subunit rRNA phylogenies of peritrichs are congruent and do not support the clustering of mobilids and sessilids (Ciliophora, Oligohymenophorea). J Eukaryot Microbiol 57:265–272
Gong Y, Yu Y, Villalobo E, Zhu F, Miao W (2006) Reevaluation of the phylogenetic relationship between mobilid and sessilid peritrichs (Ciliophora, Oligohymenophorea) based on small subunit rRNA genes sequences. J Eukaryot Microbiol 53:397–403
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Klein BM (1958) The dry silver method and its proper use. J Parasitol 5:99–103
Li H, Zhao Y (2009) Study on four species of ectoparasitic trichodinids from cultured freshwater fishes in Yibin, Sichuan Province. J Hebei Agric Sci 13:53–56 (in Chinese with english summary)
Liu C, Zhao Y (2010) Morphological and taxonomic study on four species of trichodinids parasitic on gills of Siluriformes fishes from freshwater. J Chongqing Normal Univ 27:1–5 (in Chinese with English summary)
Lom J, Hoffman GL (1964) Geographic distribution of some species of trichodinids (Ciliata: Peritricha) parasitic on fishes. J Protozool 50:30–35
Lom J (1958) A contribution to the systematics and morphology of endopasitic trichodinids from amphibians of uniform specific characteristics. J Protozool 5:251–263
Lom J (1970) Observations on trichodinid ciliates from freshwater fishes. Arch Protistenk 112:153–177
Lynn DH (2008) The ciliated protozoa: characterization, classification, and guide to the literature, 3rd edn. Springer, Dordrecht, pp 428–435
Medlin L, Elwood H, Stickel S, Sogin M (1988) The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499
Ronquist F, Huelsenbeck J (2003) MRBAYES 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Silvestri LG, Hill LG (1965) Agreement between deoxyribonucleic acid base composition and taxometric classification of gram-positive cocci. J Bacteriol 90:136–140
Swofford D (2003) PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods), version 4.0b10. Sinauer, Sunderland
Tang FH, Zhao YJ (2007) Taxonomic study on three species of Trichodina Ehrenberg, 1838 with pathologic research on gill tissue of Carassius auratus caused by Trichodina heterodentata Duncan, 1977. A study on trichodinids from freshwater fishes in Chongqing. J Chongqing Normal Univ 24:8–14
Tang FH, Zhao YJ (2010) Taxonomic study on trichodinids parasitic on gills of freshwater fish, Carassius auratus from Chongqing, China, with the description of Trichodina brevicirra sp. nov. Acta Hydrobiol Sin 34:1004–1011
Tang FH, Zhao YJ (2011) Study of trichodinids (Protozoa, Ciliophora) parasitic on gills of freshwater fishes from Chongqing, China, and identification of a new species, Trichodina cyprinocola sp. nov. Afr J Microbiol Res 5:5523–5527
Tang FH, Zhao YJ (2012a) Two trichodinids of Paratrichodina Lom, 1963 (Ciliophora, Peritrichida, Trichodinidae) infecting gills of Ietalurus punetaus from Chongqing, China. Afr J Microbiol Res 6:2145–2149
Tang FH, Zhao YJ (2012b) Record of three new Trichodina species (Protozoa, Ciliophora) parasitic on gills of freshwater fishes from Chongqing, China. Afr J Microbiol Res 6:7108–7116. doi:10.5897/AJMR12.1163
Tang FH, Zhao YJ, Chen H (2005) Trichodinid ectoparasites from golden carp, with a description of Trichodina paranigra sp. nov. Acta Hydrobiol Sin 29:75–80 (in Chinese with english summary)
Tang FH, Zhao YJ, Tang AK (2005) Presence of ectoparasitic trichodinids (Ciliophora, Oligohymenophorea, Peritrichida) on the gills of cultured freshwater fish, Carassius auratus in Chongqing, China, with the description of a new species of the genus Trichodina. Acta Zootax Sin 30:35–40
Tang FH, Zhao YJ, Tao YF (2007) Trichodinids (Ciliophora: Peritrichida) parasitic on gills of freshwater fishes, Carassius auratus and Aristichthys nobilis from China, with the description of Trichodina subtilihamata sp. nov. Zootaxa 1582:39–48
Tao YF, Zhao YJ (2006) Ectoparasitic trichodinids (Protozoa, Ciliphora, Peritrichida) from some freshwater fishes in the Chongqing area, China, with description of a new species of the genus Trichodina Ehrenberg, 1838. Acta Zootax Sin 31:784–789
Tao YF, Zhao YJ, Tang FH (2008) Seven species of trichodinid ectoparasites (Ciliophora: Peritrichida) from freshwater fishes, Hypophthalmichthys molitrix, Aristichthys nobilis and Ctenopharyngodon idellus, with the description of Trichodina chongqingensis sp. nov. Acta Hydrobiol Sin 32(suppl):124–129 (in Chinese with english summary)
Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Utz LRP, Eizirik E (2007) Molecular phylogenetics of subclass Peritrichia (Ciliophora: Oligohymenophorea) based on expanded analyses of 18S rRNA sequences. J Eukaryot Microbiol 54:303–305
Wierzbicka J (1998) Developmental stages of protozoans Trichodina kupermani Arthur et Lom, 1984 (Ciliophora, Peritrichia) parasites of blue bream, Abramis ballerus (L.). Acta Ichthyol Piscat 28:43–47
Xu KD, Song WB (2003) Distribution and revised checklist of the trichodinid ciliates of the world. In: Song W, Zhao Y, Xu K, Hu X, Gong J (eds) Pathogenic Protozoa in mariculture. Science, Beijing, pp 429–483 (in Chinese)
Yu SS, Zhao YJ, Tang FH (2011) Geographical distribution and diversity of trichodinid ectoparasites (Ciliophora, Oligohymenophorea, Mobilia) from the gills of fresh and estuarine fishes in Zhejiang Province, China and coastal regions of the East China Sea. Eur J Sci Res 64:61–74
Zhan ZF, Xu KD, Warren A, Gong YC (2009) Reconsideration of phylogenetic relationships of the subclass Peritrichia (Ciliophora, Oligohymenophorea) based on small subunit ribosomal RNA gene sequences, with the establishment of a new subclass Mobilia Kahl, 1933. J Eukaryot Microbiol 56:552–558
Zhang H, Huang Q, Du G, Wu P, Liu X (2003) Using nuclear ribosomal DNA internal transcribed spacers (ITS) to discuss the phylogeny of Thuidiaceae (S.L., Musci). Acta Botanica Yunnanica 25:491–496
Zhao YJ, Tang FH (2007) Trichodinid ectoparasites from the freshwater fish Misgurnus anguillicaudatus (Cantor) and mollusc Anodonta woodiana (Lea) of Chongqing in China, with descriptions of two new species of Trichodina Ehrenberg, 1838. Syst Parasitol 67:65–72
Zhao YJ, Tang FH (2011) Taxonomic study on trichodinids (Protozoa, Ciliophora) infecting on gills of freshwater fishes, Cyprinus carpio and Mylopharyngodon piceus from China, with the description of Trichodina regularis sp. nov. Eur J Sci Res 58:231–237
Zhao YJ, Tang FH, Tang AK (2007) A taxonomic study on species of Trichodinella (Raabe, 1950) Sramek-Husek, 1953 and Tripartiella Lom, 1959 with seasonal population dynamics of Trichodinella epizootica (Raabe, 1950) Sramek-Husek, 1953. A study on trichodinids from freshwater fishes in Chongqing. J Chongqing Normal Univ 24:1–6
Zhou Y, Zhao YJ, Tang FH (2008) Study on morphology taxonomy of epizoic trichodinid of loaches in Chongqing area. Prog Modern Biomed 28:1677–1680
Acknowledgments
This study was supported by Grants from the National Natural Science Foundation of China (Nos. 30970329, 31101637, 31172068), Project of Chongqing Science and Technology Commission (No. CSTC, 2010CA1010), the Science Research Foundation of the Education Committee of Chongqing (no. KJ090814) and Science Founding of Chongqing Normal University (No. 11XLB025).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Tang, FH., Zhao, YJ. & Warren, A. Phylogenetic Analyses of Trichodinids (Ciliophora, Oligohymenophora) Inferred from 18S rRNA Gene Sequence Data. Curr Microbiol 66, 306–313 (2013). https://doi.org/10.1007/s00284-012-0274-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-012-0274-5