Abstract
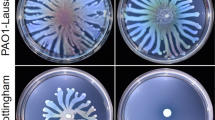
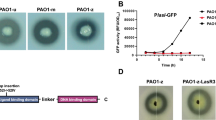
In Pseudomonas aeruginosa PAO1, the pvdQ gene has been shown to have at least two functions. It encodes the acylase enzyme and hydrolyzes 3-oxo-C12-HSL, the key signaling molecule of quorum sensing system. In addition, pvdQ is involved in swarming motility. It is required for up-regulated during swarming motility, which is triggered by high cell densities. As high-density bacterial populations also display elevated antibiotic resistance, studies have demonstrated that swarm-cell differentiation in P. aeruginosa promotes increased resistance to various antibiotics. PvdQ acts as a signal during swarm-cell differentiation, and thus may play a role in P. aeruginosa antibiotic resistance. The aim of this study is to examine whether pvdQ was involved in modifying antibiotic susceptibility during swarming conditions, and to investigate the mechanism by which this occurred. We constructed the PAO1pMEpvdQ strain, which overproduced PvdQ. PAO1pMEpvdQ promotes swarming motility, while PAO1ΔpvdQ abolishes swarming motility. In addition, both PAO1 and PAO1pMEpvdQ acquired resistance to ceftazidime, ciprofloxacin, meropenem, polymyxin B, and gentamicin, though PAO1pMEpvdQ exhibited a two to eightfold increase in antibiotic resistance compared to PAO1. These results indicate that pvdQ plays an important role in elevating antibiotic resistance via swarm-cell differentiation and possibly other mechanisms as well. We analyzed outer membrane permeability. Our data also suggest that pvdQ decreases P. aeruginosa outer membrane permeability, thereby elevating antibiotic resistance under swarming conditions. Our results suggest new approaches for reducing P. aeruginosa resistance.






Similar content being viewed by others
References
Hutchison ML, Govan JR (1999) Pathogenicity of microbes associated with cystic fibrosis. Microbes Infect 1:1005–1014
Iglewski BH, Van Delden C (1998) Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis 4:551–560
Hancock RE, Speert DP (2000) Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updat 3:247–255
Strateva T, Yordanov D (2009) Pseudomonas aeruginosa—a phenomenon of bacterial resistance. J Med Microbiol 58:1133–1148
Arora SK, Neely AN, Blair B, Lory S, Ramphal R (2005) Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect Immun 73:4395–4398
Yeung AT, Torfs EC, Jamshidi F, Bains M, Wiegand I, Hancock RE, Overhage J (2009) Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol 191:5592–5602
Rashid MH, Kornberg A (2000) Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 97:4885–4890
Harshey RM, Matsuyama T (1994) Dimorphic transition in Escherichia coli and Salmonella typhimurium: Surface-induced differentiation into hyperflagellate swarmer cells. Proc Natl Acad Sci USA 91:8631–8635
Caiazza NC, Merritt JH, Brothers KM, O’Toole GA (2007) Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:3603–3612
Fraser GM, Hughes C (1999) Swarming motility. Curr Opin Microbiol 2:630–635
Butler MT, Wang Q, Harshey RM (2010) Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci USA 107:3776–3781
Surette MG, Kim W (2003) Swarming populations of Salmonella represent a unique physiological state coupled to multiple mechanisms of antibiotic resistance. Biol Proced Online 5:189–196
Sio CF, Otten LG, Cool RH, Diggle SP, Quax WJ (2006) Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun 74:1673–1682
Ye L, Li G, Li H, Wang L, Mao Y, Song J (2011) Pseudomonas aeruginosa pvdQ gene prevents Caco-2 cells from obstruction of quorum-sensing signal. Curr Microbiol 62:32–37
Jimenez PN, Koch G, Papaioannou E, Quax WJ (2010) Role of PvdQ in Pseudomonas aeruginosa virulence under iron-limiting conditions. Microbiology 156:49–59
Lamont IL, Martin LW (2003) Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 149:833–842
Overhage J, Bains M, Brazas MD, Hancock RE (2008) Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190:2671–2679
Köhler T, Curty LK, Barja F, van Delden C, Pechère JC (2000) Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996
Maniatis T, Fritsch E, Sambrook J (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Press, Plainview
Huang JJ, Han JI, Zhang LH, Leadbetter JR (2003) Utilization of acyl-homoserine lactone quorum signals for growth by a Soil Pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 69:5941–5949
Yoneda CH, Murata T, Gotoh N, Yamamoto H, Fujiwara H, Nishino T, Shimizu E (2005) Measurement of Pseudomonas aeruginosa multidrug efflux pumps by quantitative real-time polymerase chain reaction. FEMS Microbiol Lett 243:125–131
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ΔΔCT method. Methods 25:402–408
Caiazza NC, Shanks RM, O’Toole GA (2005) Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361
Overhage J, Lewenza S, Marr AK, Hancock RE (2007) Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J Bacteriol 189:2164–2169
Peeters E, Nelis HJ, Coenye T (2008) Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods 72:157–165
Macia MD, Borrell N, Perez JL, Oliver A (2004) Detection and susceptibility testing of hypermutable Pseudomonas aeruginosa strains with the E test and disk diffusion. Antimicrob Agents Chemother 48:2665–2672
Ozhak-Baysan B, Ongut G, Oqunc D, Gunseren F, Sepin-Ozen N, Ozturk F, Aktepe OC, Gultekin M (2010) Evaluation of in vitro activities of tigecycline and various antibiotics against Brucella spp. Pol J Microbiol 59:55–60
Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE (2008) Induction by cationic antimicrobial peptides and involvement in intrinsic polymyxin and antimicrobial peptide resistance, biofilm formation, and swarming motility of PsrA in Pseudomonas aeruginosa. J Bacteriol 190:5624–5634
Loh B, Grant C, Hancock RE (1984) Use of the fluorescent probe 1-N-Phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother 26:546–551
Hocquet D, Bertrand X, Kohler T, Talon D, Plesiat P (2003) Genetic and phenotypic variations of a resistant Pseudomonas aeruginosa epidemic clone. Antimicrob Agents Chemother 47:1887–1894
Delcour AH (2009) Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–816
Bassler BL (2002) Small talk. Cell-to-cell communication in bacteria. Cell 109:421–424
Fuqua C, Parsek MR, Greenberg EP (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35:439–468
Reimmann C, Ginet N, Michel L, Keel C, Michaux P, Haas D (2002) Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology 148:923–932
Merritt JH, Brothers KM, Kuchma SL, O’Toole GA (2007) SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol 189:8154–8164
Parsek MR, Greenberg EP (2005) Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13:27–33
Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298
Cornelis P (2010) Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol 86:1637–1645
Hirsch EB, Tam VH (2010) Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res 10:441–451
Acknowledgment
This study was supported by National Natural Science Foundation of China (30873189), China National 973 project (MOST, 2007CB512900).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, L., Zhang, C., Gong, F. et al. Influence of Pseudomonas aeruginosa pvdQ Gene on Altering Antibiotic Susceptibility Under Swarming Conditions. Curr Microbiol 63, 377 (2011). https://doi.org/10.1007/s00284-011-9979-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-011-9979-0