Abstract
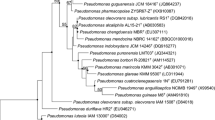
A Gram-stain-negative, non-motile, rod-shaped bacterial strain, JW-64-1T, capable of degrading methamidophos was isolated from a methamidophos-manufacturing factory in China, and was subjected to a polyphasic taxonomic investigation. Strain JW-64-1T produced circular, smooth, transparent, yellow-colored colonies (1.0–2.0 mm) on LB agar after 2 days incubation. It grew optimally at 25–30°C and pH 7.0 without the presence of NaCl. The G+C content of the total DNA was 63.6 mol%. A phylogenetic analysis based on 16S rRNA gene sequences showed that strain JW-64-1T fell within the cluster comprising Luteibacter species. The 16S rRNA gene sequence of strain JW-64-1T was most closely related to Luteibacter rhizovicinus DSM 16549T (98.6%), followed by Luteibacter yeojuensis DSM 17673T (98.4%) and L. anthropi CCUG 25036T (98.2%). The major cellular fatty acids of strain JW-64-1T were iso-C15:0 (24.1%), iso-C17:0 (20.2%) and summed feature 9 comprising iso-C17:1 ω9c and/or C16:0 10-methyl (20.3%). The major isoprenoid quinine was Q-8 (98%), and the major polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphoaminolipid, aminolipids-1, aminolipids-2, and phospholipids. The values for DNA–DNA relatedness between strain JW-64-1T and the closest phylogenetic relatives of L. rhizovicinus and Luteibacter yeojuensis were 34.8 ± 2.6 and 25.6 ± 3.1%, respectively. On the basis of the phenotypic, chemotaxonomic, DNA–DNA relatedness and phylogenetic analysis based on the 16S rRNA gene sequences, strain JW-64-1T represents a novel species of the genus Luteibacter, for which the name Luteibacter jiangsuensis sp. nov. is proposed. The type strain is JW-64-1T (=CGMCC 1.10133T = DSM 22396T).



Similar content being viewed by others
References
Breznak JA, Costilow RN (1994) Physicochemical factors in growth. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 137–154
Buck JD (1982) Nonstaining (KOH) method for determination of Gramreactions of marine bacteria. Appl Environ Microbiol 44:992–993
Chase AR, Stall RE, Hodge NC, Jones JB (1992) Characterization of Xanthomonas campestris strains from aroids using physiological, pathological, and fatty acid analysis. Phytopathology 82:754–759
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Johansen JE, Binnerup SJ, Kroer N, Mølbak L (2005) Luteibacter rhizovicinus gen. nov., sp. nov., a yellow-pigmented gammaproteobacterium isolated from the rhizosphere of barley (Hordeum vulgare L.). Int J Syst Evol Microbiol 55:2285–2291
Jung HM, Ten LN, Kim KH, An DS, Im WT, Lee ST (2009) Dyella ginsengisoli sp. nov., isolated from soil of a ginseng field in South Korea. Int J Syst Evol Microbiol 59:460–465
Kämpfer P, Lodders N, Falsen E et al (2009) Luteibacter anthropi sp. nov., isolated from human blood, and reclassification of Dyella yeojuensis as Luteibacter yeojuensis Kim et al. 2006 comb. nov. Int J Syst Evol Microbiol 59:2884–2887
Kim BY, Weon HY, Lee KH, Seok SJ, Kwon SW, Go SJ, Stackebrandt E (2006) Dyella yeojuensis sp. nov., isolated from greenhouse soil in Korea. Int J Syst Evol Microbiol 56:2079–2082
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Kumar S, Tamura K, Nei M (2004) MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Mandel M, Marmur J (1968) Use of ultraviolet absorbance-temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol 12B:195–206
Mergaert J, Cnockaert MC, Swings J (2002) Fulvimonas soli gen. nov., sp. nov., a γ-proteobacterium isolated from soil after enrichment on acetylated starch plastic. Int J Syst Evol Microbiol 52:1285–1289
Moore DD, Dowhan D (1995) Preparation and analysis of DNA. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. Wiley, New York, pp 2–11
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Swings J, Gillis M, Kersters K, De Vos P, Gossele′ F, De Ley J (1980) Frateuria, a new genus for ‘‘Acetobacter aurantius’’. Int J Syst Bacteriol 30:547–556
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202
Tindall BJ (1990) A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol 13:128–130
Wang L, Wen Y, Guo XQ, Wang GL, Li SP, Jiang JD (2010) Degradation of methamidophos by Hyphomicrobium species MAP-1 and the biochemical degradation pathway. Biodegradation 21:513–523
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Weon HY, Anandham R, Kim BY, Hong SB, Jeon YA, Kwon SW (2009) Dyella soli sp. nov. and Dyella terrae sp. nov., isolated from soils in Jeju Island, Korea. Int J Syst Evol Microbiol 59:1685–1690
Acknowledgments
We are grateful to Dr. B. J. Tindall of DSMZ, Braunschweig, Germany, for the analysis of respiratory quinones and polar lipids. This work was supported by grants from the National Programs for High Technology Research and Development of China (2007AA10Z405), the Major Projects on Control and Rectification of Water Body Pollution (2009ZX07103-002, 2008ZX07316-002) and Science & Technology Program of Ministry of Construction of People’s Republic of China (2009-K4-3).
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain JW-64-1T is FJ848571.
L. Wang and G. Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wang, L., Wang, Gl., Li, Sp. et al. Luteibacter jiangsuensis sp. nov.: A Methamidophos-Degrading Bacterium Isolated from a Methamidophos-Manufacturing Factory. Curr Microbiol 62, 289–295 (2011). https://doi.org/10.1007/s00284-010-9707-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9707-1