Abstract
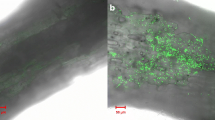
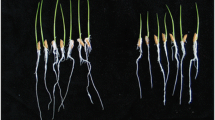
The early stage of plant–rhizobacteria interaction, affected by plant root exudates and plant–rhizobacteria surface contact, is considered to be critical for plant growth-promoting rhizobacteria colonizing plant roots and initiating the beneficial effects on plant growth. However, little is known about the mechanisms of plant–rhizobacteria surface contact involved in early stage of plant–rhizobacteria interaction. In order to reveal the molecular mechanisms of the surface contact, a rhizobacterium Bacillus amyloliquefaciens B55 was interacted with plant roots of rice R109 and used to perform a cDNA-based suppression-subtractive hybridization. Seven differentially expressed DNA fragments were identified. Except for the two fragments showing no matches to any known sequences in the Genbank, the other five fragments were found to have high homologies with the genes encoding 2-oxoglutarate dehydrogenase E1 component OdhA, aspartate ammonia-lyase AnsB, and hypothetical protein proposed to be involved in surface adhesion, acetolactate decarboxylase AlsD, and DNA mismatch repair protein MutL, respectively. The induced RNA expression levels of two putative genes ansB and odhA and an unmatched DNA fragment BD33 were verified by RT-PCR analysis.



Similar content being viewed by others
References
Carlsson P, Hederstedt L (1989) Genetic characterization of Bacillus subtilis odhA and odhB, encoding 2-oxoglutarate dehydrogenase and dihydrolipoamide transsuccinylase, respectively. J Bacteriol 171:3667–3672
Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva O, Junge H, Voigt B, Jungblut PR, Vater J, Sussmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R (2007) Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol 25:1007–1014
Chin-A-Woeng TFC, Bloemberg GV, Mulders IHM, Dekkers LC, Lugtenberg BJJ (2000) Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol Plant Microbe Interact 13:1340–1345
De Long SK, Kinney KA, Kirisits MJ (2008) Prokaryotic suppression subtractive hybridization PCR cDNA subtraction, a targeted method to identify differentially expressed genes. Appl Environ Microbiol 74:225–232
de Weert S, Vermeiren H, Mulders IHM, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, De Mot R, Lugtenberg BJJ (2002) Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant Microbe Interact 15:1173–1180
Hartmann A, Schmid M, Tuinen D, Berg G (2009) Plant-driven selection of microbes. Plant Soil 321:235–257
Kloepper JW, Hume DJ, Scher FM, Singleton C, Tipping B, Laliberte M, Frauley K, Kutchaw T, Simonson C, Lifshitz R (1988) Plant growth-promoting rhizobacteria on canola (rapeseed). Plant Dis 72:42–46
Kloepper JW, Ryu CM, Zhang S (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266
Kohler J, Hernandez JA, Caravaca F, Roldan A (2009) Induction of antioxidant enzymes is involved in the greater effectiveness of a PGPR versus AM fungi with respect to increasing the tolerance of lettuce to severe salt stress. Environ Exp Bot 65:245–252
Liu X, Bimerew M, Ma Y, Muller H, Ovadis M, Eberl L, Berg G, Chernin L (2007) Quorum-sensing signaling is required for production of the antibiotic pyrrolnitrin in a rhizospheric biocontrol strain of Serratia plymuthica. FEMS Microbiol Lett 270:299–305
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Mei JM, Nourbakhsh F, Ford CW, Holden DW (1997) Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol 26:399–407
Mohan M, Ryder S, Claypool PL, Geisert RD, Malayer JR (2002) Analysis of gene expression in the bovine blastocyst produced in vitro using suppression-subtractive hybridization. Biol Reprod 67:447–453
Neu TR (1996) Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol Rev 60:151–166
Rosenberg M, Kjelleberg S (1986) Hydrophobic interactions: role in bacterial adhesion. Adv Microb Ecol 9:353–393
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schuhegger R, Ihring A, Gantner S, Bahnweg G, Knappe C, Vogg G, Hutzler P, Schmid M, Van Breusegem F, Eberl L (2006) Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ 29:909–918
Shih IL, Van YT (2001) The production of poly-(γ-glutamic acid) from microorganisms and its various applications. Bioresour Technol 79:207–225
Somers E, Vanderleyden J, Srinivasan M (2004) Rhizosphere bacterial signalling: a love parade beneath our feet. Crit Rev Microbiol 30:205–240
Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM (2002) Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun 70:6373–6382
Steindler L, Bertani I, De Sordi L, Schwager S, Eberl L, Venturi V (2009) LasI/R and RhlI/R quorum sensing in a plant beneficial strain of Pseudomonas aeruginosa. Appl Environ Microbiol 75:5131–5140
Walker TS, Bais HP, Grotewold E, Vivanco JM (2003) Root exudation and rhizosphere biology. Plant Physiol 132:44–51
Wu H, Wang S, Qiao J, Liu J, Zhan J, Gao X (2009) Expression of HpaGXooc protein in Bacillus subtilis and its biological functions. J Microbiol Biotechnol 19:194–203
Zhang X, Miao W, Zhu G, Wang J (1993) Screening of bacteria antagonits against several important crop disease pathogens. Chin J Biol Control 9:126–129
Acknowledgments
This study was supported by grants from National Natural Science Fund of China (30570041), the National 863 Program of China (2006AA10Z172; 2006AA10A203), the Special Nonprofit Scientific Research Program, P. R. China (3–32), Program of International Science and Technology Cooperation (2009DFA32740), the Specialized Research Fund for the Doctoral Program of Higher Education, P. R. China (20060307012), and 530 program of Wuxi City Government. Dr. Ting Zhou (Guelph Food Research Center, Agriculture and Agri-Food Canada) is thanked for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, J., He, D., Ma, X. et al. Identification of Up-regulated Genes of Bacillus amyloliquefaciens B55 During the Early Stage of Direct Surface Contact with Rice R109 Root. Curr Microbiol 62, 267–272 (2011). https://doi.org/10.1007/s00284-010-9701-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9701-7