Abstract
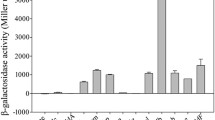
Genetic engineering of lactic acid bacteria (LAB) requires a reliable gene expression system. Especially, a stable promoter is an important genetic element to induce gene expression in such a system. We report on a novel tuf promoter (Ptuf) of Lactococcus lactis subsp. lactis IL1403 that was screened and selected through analysis of previously published microarray data. Ptuf activity was examined and compared with three other known lactococcal promoters (PdnaJ, PpfkA, and Pusp45) using different bacteria as expression hosts. Each promoter was, respectively, fused to the promoterless and modified bmpB gene as a reporter, and we estimated promoter activity through BmpB expression. All promoters were active in IL1403, and Ptuf activity was strongest among them. The activity of each promoter differed by host bacteria (Lactobacillus plantarum Lb25, Lactobacillus reuteri ATCC23272, and Escherichia coli Top10F’). Ptuf had the highest activity in IL1403 when growth reached late log phase. The activity of each promoter correlated with the expression of each cognate gene in the microarray data (R 2 = 0.7186, P = 0.06968). This study revealed that novel food-grade promoters such as IL1403 Ptuf can be selected from microarray data for food-grade microorganisms and Ptuf can be used to develop a reliable gene expression system in L. lactis.





Similar content being viewed by others
References
Alegre MT, Rodríguez MC, Mesas JM (2004) Transformation of Lactobacillus plantarum by electroporation with in vitro modified plasmid DNA. FEMS Microbiol Lett 241:73–77
Bermúdez-Humarán LG (2009) Lactococcus lactis as a live vector for mucosal delivery of therapeutic proteins. Hum Vaccin 5:4
Bron PA, Hoffer SM, Van Swam II et al (2004) Selection and characterization of conditionally active promoters in Lactobacillus plantarum, using alanine racemase as a promoter probe. Appl Environ Microbiol 70:310–317
Chen Y-S, Steele JL (2005) Analysis of promoter sequences from Lactobacillus helveticus CNRZ32 and their activity in other lactic acid bacteria. J Appl Microbiol 98:64–72
Cohen DPA, Renes J, Bouwman FG et al (2006) Proteomic analysis of log to stationary growth phase Lactobacillus plantarum cells and a 2-DE database. Proteomics 6:6485–6493
Djordjevic G, Klaenhammer T (1998) Inducible gene expression systems in Lactococcus lactis. Mol Biotechnol 9:127–139
Guillot A, Gitton C, Anglade P et al (2003) Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics 3:337–354
Jeong DW, Chul Choi Y, Min Lee J et al (2006) Isolation and characterization of promoters from Lactococcus lactis ssp. cremoris LM0230. Food Microbiol 23:82–89
Kim SH (2008) Effects of a M cell targeting peptide, CKSTHPLSC, on antigen delivery in small intestine using BmpB as fusion model antigen. Department of Agricultural Biotechnology, Seoul National University
La T, Phillips ND, Reichel MP et al (2004) Protection of pigs from swine dysentery by vaccination with recombinant BmpB, a 29.7 kDa outer-membrane lipoprotein of Brachyspira hyodysenteriae. Vet Microbiol 102:97–109
Liu C-Q, Su P, Khunajakr N et al (2005) Development of food-grade cloning and expression vectors for Lactococcus lactis. J Appl Microbiol 98:127–135
Morelli L, Vogensen FK, and Wright AV (2004) Genetics of lactic acid bacteria. In: Lactic acid bacteria: microbiological and functional aspects, 3rd edn. Marcel Dekker, NY, USA, pp 249–294
Morello E, Bermúdez-Humarán LG, Llull D et al (2008) Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J Mol Microbiol Biotechnol 14:48–58
Beyer NH, Roepstorff P, Hammer K et al (2003) Proteome analysis of the purine stimulon from Lactococcus lactis. Proteomics 3:786–797
Nouaille S, Ribeiro LA, Miyoshi A et al (2003) Heterologous protein production and delivery systems for Lactococcus lactis. Genet Mol Res 2:102–111
O’Sullivan DJ, Klaenhammer TR (1993) Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol 59:2730–2733
Redon E, Loubiere P, Cocaign-Bousquet M (2005) Role of mRNA stability during genome-wide adaptation of Lactococcus lactis to carbon starvation. J Biol Chem 280:36380–36385
Shen L, Shi Y, Douglas AL et al (2000) Identification and characterization of promoters regulating tuf expression in Chlamydia trachomatis serovar F. Arch Biochem Biophys 379:46–56
Simon D, Chopin A (1988) Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559–566
Solem C, Jensen PR (2002) Modulation of gene expression made easy. Appl Environ Microbiol 68:2397–2403
Steidler L, Rottiers P (2006) Therapeutic drug delivery by genetically modified Lactococcus lactis. Ann NY Acad Sci 1072:176–186
Team RDC (2008) R: A language and environment for statistical computing
Teuber M, Geis A (2006) The genus Lactococcus. In: The prokaryotes. Springer, NY, pp 205–228
van Asseldonk M, de Vos WM, Simons G (1993) Functional analysis of the Lactococcus lactis usp45 secretion signal in the secretion of a homologous proteinase and a heterologous alpha-amylase. Mol Gen Genet 240:428–434
Ventura M, Canchaya C, Meylan V et al (2003) Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl Environ Microbiol 69:6908–6922
Wells JM, Mercenier A (2008) Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat Rev Microbiol 6:349–362
Yun JH, Lee KB, Sung YK et al (2008) Isolation and characterization of potential probiotic lactobacilli from pig feces. J Basic Microbiol 48:1–7
Acknowledgments
This study was supported by a graduate fellowship from the BK 21 project and the Research Institute for Agriculture and Life Sciences, Seoul National University. We thank Dr. Alain Chopin (Laboratoire de Génétique Microbienne, France) and Takashi Sasaki (Meiji Dairies Corporation, Japan) for providing the pIL252 vector and the IL1403 strain.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, E.B., Piao, D.C., Son, J.S. et al. Cloning and Characterization of a Novel tuf Promoter from Lactococcus lactis subsp. lactis IL1403. Curr Microbiol 59, 425–431 (2009). https://doi.org/10.1007/s00284-009-9455-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-009-9455-2