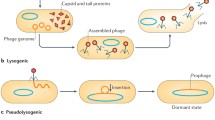
Abstract
We show by electron microscopy that Lactobacillus gasseri phage LgaI, a temperate phage residing in the chromosome of Lactobacillus gasseri ATCC33323, belongs to the family of Myoviridae phages. The LgaI DNA is packed by the “head-full” mechanism, as demonstrated by analysis of restriction patterns of heated (74°C) or non-heated DNA. By isolating prophage-cured cells, we were able to demonstrate phage LgaI to be responsible for the strong autolytic phenotype observed for Lactobacillus gasseri ATCC33323. In addition, we show that a copy of the LgaI prophage resides in the chromosome of Lactobacillus gasseri NCK102. The LgaI prophage was not inducible in L. gasseri NCK102-adh by mitomycin C, however, it apparently contributed to the autolytic phenotype of this strain.




Similar content being viewed by others
References
Altermann E, Klein JR, Henrich B (1999) Primary structure and features of the genome of the Lactobacillus gasseri temperate bacteriophage φadh. Gene 236:333–346
Azcarate-Peril MA, Altermann E, Goh YJ, Tallon R, Sanozky-Dawes RB, Pfeiler E, O’Flaherty S, Buck BL, Dobson A, Duong T, Miller MJ, Barrangou R, Klaenhammer TR (2008) Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl Environ Microbiol 74:4610–4625
Brüssow H, Desiere F (2001) Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol 39:213–222
de Man JC, Rogosa M, Sharpe ME (1960) A medium for the cultivation of Lactobacilli. J Appl Bact 23:130–135
Engel G, Altermann E, Klein JR, Henrich B (1998) Structure of a genome region of the Lactobacillus gasseri temperate phage φadh covering a repressor gene and cognate promoters. Gene 210:61–70
Husson-Kao C, Mengaud J, Cesselin B, van Sinderen D, Benbadis L, Chapot-Chartier MP (2000) The Streptococcus thermophilus autolytic phenotype results from a leaky prophage. Appl Environ Microbiol 66:558–562
Kilic AO, Pavlova SI, Alpay S, Kilic SS, Tao L (2001) Comparative study of vaginal Lactobacillus phages isolated from women in the United States and Turkey: prevalence, morphology, host range, and DNA homology. Clin Diagn Lab Immunol 8:31–39
Kleeman EG, Klaenhammer TR (1982) Adherence of Lactobacillus species to human fetal intestinal cells. J Dairy Sci 65:2063–2069
Lauer E, Helming C, Kandler O (1980) Heterogeneity of the species Lactobacillus acidophilus (Moro) Hansen and Moquot as revealed by biochemical characteristics and DNA–DNA hybridization. Zbl Bakt Hyg I Abt Orig C 1:150–168
Leenhouts KJ, Kok J, Venema G (1990) Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl Environ Microbiol 56:2726–2735
Mahony J, McGrath S, Fitzgerald GF, van Sinderen D (2008) Identification and characterization of lactococcal-prophage-carried superinfection exclusion genes. Appl Environ Microbiol 74:6206–6215
Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavolv A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goldstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee L-H, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O’Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D (2006) Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA 103:15611–15616
McGrath S, Fitzgerald GF, van Sinderen D (2002) Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol Microbiol 43:509–520
Raya RR, Fremaux C, De Antoni GL, Klaenhammer TR (1992) Site-specific integration of the temperate bacteriophage φadh into the Lactobacillus gasseri chromosome and molecular characterization of the phage (attP) and bacterial (attB) attachment sites. J Bacteriol 174:5584–5592
Raya RR, Kleeman EG, Luchansky JB, Klaenhammer TR (1989) Characterization of the temperate bacteriophage φadh and plasmid transduction in Lactobacillus acidophilus ADH. Appl Environ Microbiol 55:2206–2213
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Sugahara K, Yokoi KJ, Nakamura Y, Nishino T, Yamakawa A, Taketo A, Kodaira K (2007) Mutational and biochemical analyses of the endolysin LysgaY encoded by the Lactobacillus gasseri JCM 1131T phage φgaY. Gene 404:41–52
Sun X, Göhler A, Heller KJ, Neve H (2006) The ltp gene of temperate Streptococcus thermophilus phage TP-J34 encodes a lipoprotein which is expressed during lysogeny. Virology 350:146–157
Ventura M, Canchaya C, Bernini V, Altermann E, Barrangou R, McGrath S, Claesson MJ, Li Y, Leahy S, Walker CD, Zink R, Neviani E, Steele J, Broadbent J, Klaenhammer TR, Fitzgerald GF, O’Toole PW, van Sinderen D (2006) Comparative genomics and transcriptional analysis of prophages identified in the genomes of Lactobacillus gasseri, Lactobacillus salivarius, and Lactobacillus casei. Appl Environ Microbiol 72:3130–3146
Villion M, Moineau S (2009) Bacteriophages of Lactobacillus. Front Biosci 14:1661–1683
Yokoi KJ, Kawasaki K, Taketo A, Kodaira K (2004) Characterization of lytic enzyme activities of Lactobacillus gasseri with special reference to autolysis. Int J Food Microbiol 96:273–279
Yokoi KJ, Shinohara M, Kawahigashi N, Nakagawa K, Kawasaki K, Nakamura S, Taketo A, Kodaira K (2005) Molecular properties of the two-component cell lysis system encoded by prophage φgaY of Lactobacillus gasseri JCM 1131T: cloning, sequencing, and expression in Escherichia coli. Int J Food Microbiol 99:297–308
Acknowledgements
E. A. I. gratefully acknowledges funding by the Egyptian Government (Long Term Mission). We thank B. Henrich (University of Kaiserslautern, Germany) for kindly providing L. gasseri strains NCK102 and NCK102-adh, and Bernd Fahrenholz for technical help in preparing phage samples for electron microscopy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ismail, E.A., Neve, H., Geis, A. et al. Characterization of Temperate Lactobacillus gasseri Phage LgaI and Its Impact as Prophage on Autolysis of Its Lysogenic Host Strains. Curr Microbiol 58, 648–653 (2009). https://doi.org/10.1007/s00284-009-9384-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-009-9384-0