Abstract
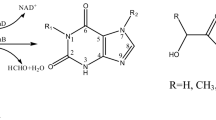
Pseudomonas sp. HK-6 is able to utilize 2,4,6-trinitrotoluene (TNT) as a sole nitrogen source. The pnrB gene of the HK-6 strain was cloned using degenerate primers synthesized on the basis of the sequence information of the terminal amino acids of a previously purified native TNT nitroreductase. The nucleotide sequence of pnrB was 654 bp long, and its deduced polypeptide sequence was composed of 217 amino acid residues with a predicted molecular mass of 24 kDa. To facilitate the purification and characterization of this enzyme, an Escherichia expression plasmid harboring six histidine residues fused to a pnrB gene was constructed (His6-PnrB) and designated pPSC1. The His6-PnrB induced in E. coli BL21 was purified using a nickel affinity column to homogeneity. Its enzymatic activity was assayed by measuring absorbance changes at 340 nm due to NADH oxidation. The Vmax and K m values of the enzyme for TNT were 12.6 μmol/min/mg protein and 2.9 mM, respectively. In addition, the pnrB knockout mutant was constructed via a single-crossover homologous recombination with a partial pnrB DNA fragment that lacked both start and stop codons. Eight days was required for complete degradation of 0.5 mM TNT by the wild-type HK-6 strain, whereas the pnrB mutant degraded only 10% of the TNT in the same time period. Even after 20 days, only approximately 50% of the 0.5 mM TNT was degraded by the pnrB mutant. These results illustrate that pnrB may perform a crucial role in the TNT degradation pathway of the HK-6 strain.



Similar content being viewed by others
References
Bryant C, DeLuca M (1991) Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacteria cloacae. J Biol Chem 266:4119–4125
Caballero A, Lazaro JJ, Ramos JL, Esteve-Nunez A (2005) PnrA, a new nitroreductase-family enzyme in the TNT-degrading strain Pseudomonas putida JRL11. Environ Microbiol 7:1211–1219
Chang HW, Kahng HY, Kim SI, Chun JW, Oh KH (2004) Characterization of Pseudomonas sp. HK-6 cells responding to explosive RDX (hexahydro-1,3,5-trinitro-1,3,5,-triazine). Appl Microbiol Biotechnol 65:323–329
Comfort SD, Shea PJ, Hundal LS, Li Z, Woodbury BL, Martin L, Powers WL (1995) TNT transport and fate in contaminated soil. J Environ Qual 24:1174–1182
Esteve-Núñez A, Caballero A, Ramos JL (2001) Biological degradation of 2,4,6-trinitrotoluene. Microbiol Mol Biol Rev 65:335–352
Esteve-Núñez A, Luchessi G, Philipps B, Schink B, Ramos JL (2000) Respiration of 2,4,6-trinitrotoluene by Pseudomonas sp. strain JLR11. J Bacteriol 182:1352–1355
French CE, Nicklin S, Bruce NC (1998) Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase. Appl Environ Microbiol 64:2864–2868
Hannink N, Rosser SJ, French CE, Basran A, Murray JA, Nicklin S, Bruce NC (2001) Phytodetoxification of TNT by transgenic plants expressing a bacterial nitroreductase. Nat Biotechnol 19:1168–1172
Ho EM, Chang HW, Kim SI, Kahng HY, Oh KH (2004) Analysis of TNT (2,4,6-trinitrotoluene)-inducible cellular responses and stress shock proteome in Stenotrophomonas sp. OK-5. Curr Microbiol 49:346–352
Kahng HY, Lee BU, Cho YS, Oh KH (2007) Purification and characterization of the NAD(P)H-nitroreductase for the catabolism of 2,4,6-trinitrotoluene (TNT) in Pseudomonas sp. HK-6. Biotechnol Bioprocess Eng 12:433–440
Klausmeier RE, Osmon JL, Walls DR (1973) The effect of trinitrotoluene on microorganisms. Dev Ind Microbiol 15:309–317
Kobori T, Sasaki H, Lee WC, Zenno S, Saigo K, Murphy ME, Tanokura M (2001) Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution. J Biol Chem 276:2816–2823
Lee MS, Chang HW, Kahng HY, So JS, Oh KH (2002) Biological removal of explosive 2,4,6-trinitrotoluene by Stenotrophomonas sp. OK-5 in bench-scale bioreactors. Biotechnol Bioprcess Eng 7:105–111
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Nelson KE, Weinel C, Paulsen IT, et al. (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4:799–808
Smock LA, Stoneburner DL, Clark JR (1976) The toxic effects of trinitrotoluene (TNT) and its primary degradation products on two species of algae and fathered minnow. Water Res 10:537–543
Spratt BG, Hedge PJ, te Heesen S, Edelman A, Broom-Smith JK (1986) Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337–342
Stemmer WP (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389–391
Van Dillewijn P, Caballero A, Paz JA, Gonzalez-Perez MM, Oliver JM, Ramos JL (2007) Bioremediation of 2,4,6-trinitrotoluene under field conditions. Environ Sci Technol 41:1378–1–1383
Zenno S, Koike H, Kumar AN, Jayaraman R, Tanokumura M, Saigo K (1996) Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol 178:4508–4514
Zenno S, Koike H, Tanokumura M, Saigo K (1996) Gene cloning, purification and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J Biochem 120:736–744
Zenno S, Saigo K, Kanoh H, Inouye S (1994) Identification of the gene encoding the major NAD(P)H-flavin oxidoreductase of the bioluminescent bacterium Vibrio fischeri ATCC 7744. J Bacteriol 176:3536–3543
Acknowledgments
This work was supported by a grant (R01-2005-000-106080) from the Basic Research Program of the Korea Science & Engineering Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, BU., Park, SC., Cho, YS. et al. Expression and Characterization of the TNT Nitroreductase of Pseudomonas sp. HK-6 in Escherichia coli . Curr Microbiol 56, 386–390 (2008). https://doi.org/10.1007/s00284-007-9093-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-007-9093-5