Abstract
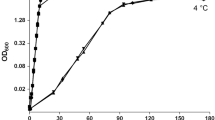
Most research on the adaptation of thermophiles is focused on their adaptation to heat stress; only a few studies are focused on their cold adaptation. In this report, the thermophilic bacterium Rhodothermus sp. XMH10 was examined to gain a better understanding of gene expression in response to low temperature. Random arbitrarily primed polymerase chain reaction (RAP-PCR) was used to isolate and identify differentially expressed genes of bacteria grown at 45°C (lowest) compared to those at 75°C (optimal). Fifty-three differential cDNA fragments in total were isolated. Among them, 35 different cDNAs were analyzed by Northern blot, and 17 were confirmed to be differentially expressed at the transcriptional levels. These genes reflected a profile of differential expression and were involved in many physiological processes such as metabolism, cell membrane alterations, and regulatory adaptive response; most of them have never been previously reported. This study provides some new information on the adaptation of thermophilic bacteria to environmental temperature stress.



Similar content being viewed by others
References
Berger F, Morellet N, Menu F, et al. (1996) Cold shock and cold acclimation proteins in the psychrotrophic bacterium Arthrobacter globiformis SI55. J Bacteriol 178:2999–3007
Bjornsdottir SH, Blondal T, Hreggvidsson GO, et al. (2006) Rhodothermus marinus: physiology and molecular biology. Extremophiles 1:1–16
Bock AK, Kunow J, Glasemacher J, et al. (1996) Catalytic properties, molecular composition and sequence alignments of pyruvate: ferredoxin oxidoreductase from the methanogenic archaeon Methanosarcina barkeri (strain Fusaro). Eur J Biochem 237:35–44
Fislage R, Berceanu M, Humboldt Y, et al. (1997) Primer design for a prokaryotic differential display RT-PCR. Nucleic Acids Res 25:1830–1835
Georgieva DN, Stoeva S, Ivanova V, et al. (2000) Specificity of a neutral Zn-dependent proteinase from Thermoactinomyces sacchari toward the oxidized insulin B chain. Curr Microbiol 41:70–72
Kambampati R, Lauhon CT (2003)MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42:1109–1117
Kevil CG, Walsh L, Laroux FS, et al. (1997) An improved, rapid northern protocol. Biochem Biophys Res Commun 238:277–279
Kletzin A, Adams MW (1996) Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol 178:248–257
Kuwahara T, Yamashita A, Hirakawa H, et al. (2004) Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci USA 101:14,919–14,924
Liu S, Graham JE, Bigelow L, et al. (2002) Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl Environ Microbiol 68:1697–1705
Lu S (1999) Current protocols for molecular biology, 2nd ed. Peking Union Medical College Press, Beijing
Maguire BA, Wild DG (1997) The roles of proteins L28 and L33 in the assembly and function of Escherichia coli ribosomes in vivo. Mol Microbiol 23:237–245
Mello BA, Tu Y (2003) Perfect and near-perfect adaptation in a model of bacterial chemotaxis. Biophys J 84:2943–2956
Menon S, Stahl M, Kumar R, et al. (1999) Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from Escherichia coli. J Biol Chem 274:26,743–26,750
Nakano MM, Zuber P (1998) Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu Rev Microbiol 52:165–190
Qing G, Ma LC, Khorchid A, et al. (2004) Cold-shock induced high-yield protein production in Escherichia coli. Nat Biotechnol 22:877–882
Rivera-marrero CA, Burroughs MA, Masse RA, et al. (1998) Identification of genes differentially expressed in Mycobacterium tuberculosis by differential display PCR. Microb Pathol 25:307–316
Ruan L, Liu X, Yang H, et al. (2006) Isolation and identification of thermophilic microorganisms. J Xiamen Univ (Nat Sci) 45:276–279 (in Chinese)
Shepard BD, Gilmore MS (1999) Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl Environ Microbiol 65:1470–1476
Thieringer HA, Jones PG, Inouye M (1998) Cold shock and adaptation. BioEssays 20:49–57
Topanurak S, Sinchaikul S, Sookkheo B, et al. (2005) Functional proteomics and correlated signaling pathway of the thermophilic bacterium Bacillus stearothermophilus TLS33 under cold-shock stress. Proteomics 5:4456–4471
Uyttendaele M, Schukkink R, Van Gemen B, et al. (1996) Influence of bacterial age and pH of lysis buffer on type of nucleic acid isolated. J Microbiol Methods 26:133–138
Walters DM, Rouviere PE (2005) High-density sampling differential display of prokaryotic mRNAs with RAP-PCR. Methods Mol Biol 317:85–98
Welsh J, Chada K, Dalal SS, et al. (1992) Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res 20:4965–4970
Yukti B, Saroj M, Bisaria VS (2002) Microbial β-glucosidases: cloning, properties, and applications. Crit Rev Botechnol 22:375–407
Acknowledgments
This work was supported by the China Ocean Mineral Resources R&D Association (DY105-02-04-05) and the Hi-Tech Research and Development Program of China (863 Program of China) (2004AA621010).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ruan, L., Luo, T., Li, F. et al. Identification of Differentially Expressed Genes from Rhodothermus sp. XMH10 in Response to Low Temperature Using Random Arbitrarily Primed PCR. Curr Microbiol 55, 543–548 (2007). https://doi.org/10.1007/s00284-007-9029-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-007-9029-0