Abstract
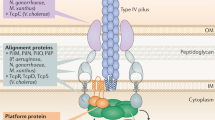
Twitching motility allows Pseudomonas aeruginosa to respond to stimuli by extending and retracting its type IV pili (TFP). PilJ is a protein necessary for this surface-associated twitching motility and bears high sequence identity with Escherichia coli methyl-accepting chemotaxis proteins (MCP). Here, we report that whereas wild-type P. aeruginosa PAO1 cells have extended pili at a single pole, pilJ mutant cells have shortened pili often at both poles despite normal levels of pilin accumulation, suggesting that PilJ is required for full TFP assembly/extension. Using yellow fluorescent protein fusions (pilJ-yfp), both plasmid born and in-frame chromosomal constructs, we determined that PilJ localizes to both poles of the cell. Overexpression of pilJ-yfp resulted in the protein accumulating between the poles.



Similar content being viewed by others
References
Alley MR, Maddock JR, Shapiro L (1992) Polar localization of a bacterial chemoreceptor. Genes Dev 6:825–836
Blackhart BD, Zusman DR (1985) “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci USA 82:8767–8770
Boyd JM (2000) Localization of the histidine kinase PilS to the poles of Pseudomonas aeruginosa and identification of a localization domain. Mol Microbiol 36:153–162
Bradley DE (1980) A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol 26:146–154
Chiang P, Habash M, Burrows LL (2005) Disparate subcellular localization patterns of Pseudomonas aeruginosa Type IV pilus ATPases involved in twitching motility. J Bacteriol 187:829–839
Croft L, Beatson SA, Whitchurch CB, et al. (2000) An interactive web-based Pseudomonas aeruginosa genome database: discovery of new genes, pathways and structures. Microbiology 146(Pt 10):2351–2364
Darzins A (1994) Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol 11:137–153
Darzins A (1993) The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single-domain response regulator CheY. J Bacteriol 175:5934–5944
Darzins A, Russell MA (1997) Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system: a review. Gene 192:109–115
Den Blaauwen T, Buddelmeijer N, Aarsman ME, et al. (1999) Timing of FtsZ assembly in Escherichia coli. J Bacteriol 181:5167–5175
Harrison DM, Skidmore J, Armitage JP, et al. (1999) Localization and environmental regulation of MCP-like proteins in Rhodobacter sphaeroides. Mol Microbiol 31:885–892
Henrichsen J (1983) Twitching motility. Annu Rev Microbiol 37:81–93
Huang B, Whitchurch CB, Mattick JS (2003) FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J Bacteriol 185:7068–7076
Maddock JR, Shapiro L (1993) Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717–1723
McMichael JC (1992) Bacterial differentiation within Moraxella bovis colonies growing at the interface of the agar medium with the Petri dish. J Gen Microbiol 138:2687–2695
Merz AJ, So M, Sheetz MP (2000) Pilus retraction powers bacterial twitching motility. Nature 407:98–102
Mignot T, Merlie JP Jr, Zusman DR (2005) Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science 310:855–857
Schweizer HP (1992) Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol 6:1195–1204
Skerker JM, Berg HC (2001) Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA 98:6901–6904
Smith AW, Iglewski BH (1989) Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res 17:10509
Sogaard-Andersen L (2004) Cell polarity, intercellular signalling and morphogenetic cell movements in Myxococcus xanthus. Curr Opin Microbiol 7:587–593
Sourjik V, Berg HC (2000) Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol 37:740–751
Stover CK, Pham XQ, Erwin AL, et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959–964
Sun H, Zusman DR, Shi W (2000) Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol 10:1143–1146
Wadhams GH, Martin AC, Armitage JP (2000) Identification and localization of a methyl-accepting chemotaxis protein in Rhodobacter sphaeroides. Mol Microbiol 36:1222–1233
Wadhams GH, Warren AV, Martin AC, Armitage JP (2003) Targeting of two signal transduction pathways to different regions of the bacterial cell. Mol Microbiol 50:763–770
Ward MJ, Zusman DR (1997) Regulation of directed motility in Myxococcus xanthus. Mol Microbiol 24:885–893
Whitchurch CB, Leech AJ, Young MD, et al. (2004) Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52:873–893
Acknowledgments
We thank Ellen Quardokus for her technical assistance and valuable discussions. This work was supported by NIH grant No. GM61318-01.
Author information
Authors and Affiliations
Corresponding author
Additional information
Paul DeLange and Tracy Collins contributed equally to this work.
Rights and permissions
About this article
Cite this article
DeLange, P.A., Collins, T.L., Pierce, G.E. et al. PilJ Localizes to Cell Poles and Is Required for Type IV Pilus Extension in Pseudomonas aeruginosa . Curr Microbiol 55, 389–395 (2007). https://doi.org/10.1007/s00284-007-9008-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-007-9008-5