Abstract
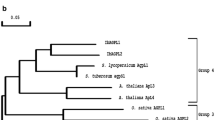
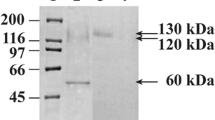
Glucoamylase obtained from Rhizopus sp. is frequently preferred for certain applications of starch modification or saccharification. The predominant enzyme, which contains a starch-binding domain on the amino terminus, has been previously characterized from several species. Additionally, the cDNA encoding this protein was cloned and found to show 100% identity to a R. oryzae strain 99-880 that has recently been sequenced by the Broad Institute of Harvard and Massachusetts Institute of Technology. Analysis of this genome indicates coding regions for two additional glucoamylase genes, amyC and amyD, lacking a starch-binding domain. The two genes encode proteins that are approximately 50% identical to the catalytic region of the AmyA protein and 67% identical to each other. The predicted AmyC and AmyD proteins contain the highly conserved signature sequences of Family 15 glycoside hydrolases. The two genes appear to be transcriptionally expressed in cultures grown in fermentable and gluconeogenic carbon sources. The predicted 49.7-kD AmyC and 48.8-kD AmyD proteins were expressed in several different ways using Pichia pastoris. When the sequence for putative secretion signal was left intact, glucoamylase activity was detected in the crude cell extracts, but no activity was present in the growth medium. However, replacement of this region with the yeast alpha-secretion signal resulted in secretion of active glucoamylase that was able to degrade soluble starch.



Similar content being viewed by others
Literature Cited
Abe A, Sone T, Sujaya IN, et al. (2003) rDNA ITS sequence of Rhizopus oryzae: its application to classification and identification of lactic acid producers. Biosci Biotechnol Biochem 67:1725–1731
Ashikari T, Nakamura N, Tanaka Y, et al. (1986) Rhizopus raw starch degrading glucoamylase: its cloning and expression in yeast. Agric Biol Chem 50:957–964
Bendtsen JD, Nielsen H, vonHeijne G, et al. (2004) Improved prediction signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Hata Y, Ishida H, Ichikawa E, et al. (1998) Nucleotide sequence of an alternative glucoamylase-encoding gene (glaB) expressed in solid-state culture of Aspergillus oryzae. Gene 207:127–134
Ishida H, Hata Y, Ichikawa E, et al. (1998) Regulation of the glucoamylase-encoding gene (glaB), expressed in solid-state culture (Koji) of Aspergillus oryzae. J Ferm Bioeng 86:301–307
Lipman DJ, Pearson WR (1985) Rapid and sensitive protein similarity searches. Science 227:1435–1441
Liu SH, Chou WI, Sheu CC, Chang MDT (2005) Improved secretory production of glucoamylase in Pichia pastoris by combination of genetic manipulations. Biochem Biophys Res Commun 326:817–824
Mertens JA, Skory CD (2007) Isolation and characterization of a second glucoamylase gene without a starch binding domain from Rhizopus oryzae. Enzyme Microbiol Tech 40:874–880
Rhizopus oryzae Sequencing Project. Broad Institute of Harvard and MIT (http://www.broad.mit.edu). Accessed August 2006
Saito K, Saito A, Ohnishi M, Oda Y (2004) Genetic diversity in Rhizopus oryzae strains as revealed by the sequence of lactate dehydrogenase genes. Arch Microbiol 182:30–36
Skory CD (2002) Homologous recombination and double strand break repair in the transformation of Rhizopus oryzae. Mol Genet Genomics 268:397–406
Takahashi T, Tsuchida Y, Irie M (1978) Purification and some properties of three forms of glucoamylase from a Rhizopus species. J Biochem 84:1183–1194
Takahashi T, Tsuchida Y, Irie M (1982) Isolation of two inactive fragments of Rhizopus sp. glucoamylase: relationship among three forms of the enzyme and the isolated fragments. J Biochem 92:1623–1633
Yu R, Hang YD (1991) Purification and characterization of a glucoamylase from Rhizopus oryzae. Food Chem 40:301–308
Author information
Authors and Affiliations
Corresponding author
Additional information
The mention of firm names or trade products does not imply that they are endorsed or recommended by the U.S. Department of Agriculture over other firms or similar products not mentioned.
Rights and permissions
About this article
Cite this article
Mertens, J.A., Skory, C.D. Isolation and Characterization of Two Genes That Encode Active Glucoamylase Without a Starch Binding Domain from Rhizopus oryzae . Curr Microbiol 54, 462–466 (2007). https://doi.org/10.1007/s00284-006-0655-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-006-0655-8