Abstract
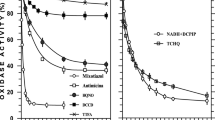
Subcellular location, chlorate specificity, and sensitivity to micromolar concentrations of azide suggest that most of the anaerobically induced nitrate reductase (NR) activity in Bradyrhizobium sp. (Lupinus) could be ascribed to the membrane type of bacterial dissimilatory NRs. Two active complexes of the enzyme, NRI of 140 kDa and NRII of 190 kDa, were detected in membranes of the nitrate-respiring USDA strain 3045. Both enzyme forms were purified to homogeneity. Obtained specific antibodies showed that these native species were immunologically closely related and composed of largely similar 126-kDa, 65-kDa, and 25-kDa subunits. The finding that NRI and NRII share common epitopes suggests that they may not be different species, but rather two forms of the same enzyme.



Similar content being viewed by others
Literature Cited
Alikulov ZA, Burikhanov SS, Sergeev NS, Lvov NP, Kretovich VL (1980) Nitratreduktaza bakteroidov klubienkov lupina. Prikl Biochim Mikrob (Transl) 16:511–516
Bedzyk L, Wang T, Ye RW (1999) The periplasmic nitrate reductase in Pseudomonas sp. strain G-179 catalyzes the first step of denitrification. J Bacteriol 181:2802–2806
Bell LC, Richardson DJ, Ferguson SJ (1990) Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. FEBS Lett 265:85–87
Berks BC, Ferguson SJ, Moir JW, Richardson DJ (1995) Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta 1232:97–173
Bhandari B, Naik MS, Nicholas DJD (1984) ATP production coupled to the denitrification of nitrate in Rhizobium japonicum, grown in cultures and in bacteroids from Glycine max. FEBS Lett 168:321–326
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bryan JK (1977) Molecular weights of protein multimers from polyacrylamide gel electrophoresis. Anal Biochem 78:513–519
Davis BJ (1964) Disc electrophoresis. II. Method and application to human serum proteins. Ann NY Acad Sci 121:404–436
Delgado MJ, Bonnard N, Tresierra-Ayala A, Bedmar EJ, Muller P (2003) The Bradyrhizobium japonicum napEDABC genes encoding the periplasmic nitrate reductase are essential for nitrate respiration. Microbiology 149:3395–3403
Delgado MJ, Fernández-López M, Bedmar EJ (1998) Soluble and membrane-bound nitrate reductase from Bradyrhizobium japonicum bacteroids. Plant Physiol Biochem 36:279–283
Fernández-López M, Olivares J, Bedmar EJ (1994) Two differentially regulated nitrate reductases required for nitrate-dependent, microaerobic growth of Bradyrhizobium japonicum. Arch Microbiol 162:310–315
Fernández-López M, Olivares J, Bedmar EJ (1996) Purification and characterization of the membrane-bound nitrate reductase isoenzymes of Bradyrhizobium japonicum. FEBS Lett 392:1–5
Galibert F, Finan TM, Long SR, et al. (2001) The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672
Kaneko T, Nakamura Y, Sato S, et al. (2002) Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA 110. DNA Res 9:189–197
Kengen SW, Rikken GB, Hagen WR, van Ginkel CG, Stams AJ (1999) Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J Bacteriol 181:6706–6711
Kennedy IR, Riguard J, Trinchant JC (1975) Nitrate reductase from bacteroids of Rhizobium japonicum: enzyme characteristic and possible interaction with nitrogen fixation. Biochim Biophys Acta 397:24–35
Kleinhofs A, Narayanan KR, Somers DA, Kuo TM, Warner RL (1986) Immunochemical methods for higher plant nitrate reductase. In: Linskens HF, Jackson JF (eds). Modern methods of plant analysis, new series, vol. 4. Berlin: Springer-Verlag, pp 190–211
Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 277:680–685
Moreno-Vivián C, Ferguson SJ (1998) Definition and distinction between assimilatory, dissimilatory and respiratory pathways. Mol Microbiol 29:664–666
Moreno-Vivián C, Cabello P, Martinez-Luque M, Blasco R, Castillo F (1999) Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol 181:6573–6584
Philippot L, Højberg O (1999) Dissimilatory nitrate reductases in bacteria. Biochim Biophys Acta 1446:1–23
Philippot L (2002) Denitrifying genes in bacterial and Archaeal genomes. Biochim Biophys Acta 1577:355–376
Polcyn W, Luciński R (2001) Functional similarities of nitrate reduction from yellow lupine bacteroids to bacterial denitrification system. J Plant Physiol 158:829–834
Polcyn W, Luciński R (2003) Aerobic and anaerobic nitrate and nitrite reduction in free-living cells of Bradyrhizobium sp. (Lupinus). FEMS Microbiol Lett 226:331–337
Richardson DJ, Berks BC, Russell DA, Spiro S, Taylor CJ (2001) Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol Life Sci 58:165–178
Weiss RL (1976) Protoplast formation in Escherichia coli. J Bacteriol 128:668–670
Zumft W (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616
Acknowledgments
This work was supported by grants from the Polish State Committee for Scientific Research: 6 P04C 026 20 to R. L. and 2 P06R 077 27 to W. P. (years 2004–2007).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Polcyn, W., Luciński, R. Dissimilatory Nitrate Reductase from Bradyrhizobium sp. (Lupinus): Subcellular Location, Catalytic Properties, and Characterization of the Active Enzyme Forms. Curr Microbiol 52, 231–237 (2006). https://doi.org/10.1007/s00284-005-0265-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-005-0265-x