Abstract
Epithelial cells, which are non-immune cells, not only function as a physical defence barrier but also continuously monitor and eliminate aberrant epithelial cells in their vicinity. In other words, it has become evident that epithelial cells possess immune cell-like functions. In fact, recent research has revealed that epithelial cells recognise the Major Histocompatibility Complex I (MHC-I) of aberrant cells as a mechanism for surveillance. This cellular defence mechanism of epithelial cells probably detects aberrant cells more promptly than the conventional immune response, making it a novel and primary biological defence. Furthermore, there is the potential for this new immune-like biological defence mechanism to establish innovative treatment for disease prevention, leading to increasing anticipation for its future medical applications. In this review, we aim to summarise the recognition and attack mechanisms of aberrant cells by epithelial cells in mammals, with a particular focus on the field of cancer. Additionally, we discuss the potential therapeutic applications of epithelial cell-based defence against cancer, including novel prophylactic treatment methods based on molecular mechanisms.





Similar content being viewed by others
Data availability
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
Change history
24 April 2024
A Correction to this paper has been published: https://doi.org/10.1007/s00281-024-01009-6
References
Morata G, Ripoll P (1975) Minutes — mutants of Drosophila autonomously affecting cell-division rate. Dev Biol 42(2):211–221. https://doi.org/10.1016/0012-1606(75)90330-9
Van Neerven SM, Vermeulen L (2023) Cell competition in development, homeostasis and cancer. Nat Rev Mol Cell Biol 24(3):221–236. https://doi.org/10.1038/s41580-022-00538-y
Maruyama T, Fujita Y (2017) Cell competition in mammals — novel homeostatic machinery for embryonic development and cancer prevention. Curr Opin Cell Biol 48:106–112. https://doi.org/10.1016/j.ceb.2017.06.007
Maruyama T, Fujita Y (2022) Cell competition in vertebrates—a key machinery for tissue homeostasis. Curr Opin Genet Dev 72:15–21. https://doi.org/10.1016/j.gde.2021.09.006
Hogan C, Dupré-Crochet S, Norman M et al (2009) Characterization of the interface between normal and transformed epithelial cells. Nat Cell Biol 11(4):460–467. https://doi.org/10.1038/ncb1853
Kajita M, Hogan C, Harris AR et al (2010) Interaction with surrounding normal epithelial cells influences signalling pathways and behaviour of Src-transformed cells. J Cell Sci 123(2):171–180. https://doi.org/10.1242/jcs.057976
Wagstaff L, Goschorska M, Kozyrska K et al (2016) Mechanical cell competition kills cells via induction of lethal p53 levels. Nat Commun 7. https://doi.org/10.1038/ncomms11373
Tamori Y, Bialucha CU, Tian AG et al (2010) Involvement of Lgl and Mahjong/VprBP in cell competition. Plos Biol 8 (7). https://doi.org/10.1371/journal.pbio.1000422
Chiba T, Ishihara E, Miyamura N, R. Narumi et al (2016) MDCK cells expressing constitutively active Yes-associated protein (YAP) undergo apical extrusion depending on neighboring cell status. Sci Rep 6. https://doi.org/10.1038/srep28383
Moya IM, Castaldo SA, Van den Mooter L et al (2019) Peritumoral activation of the Hippo pathway effectors YAP and TAZ suppresses liver cancer in mice. Science 366(6468):1029–1034. https://doi.org/10.1126/science.aaw9886
Alcolea MP, Jones PH (2015) Cell competition: winning out by losing notch. Cell Cycle 14(1):9–17. https://doi.org/10.4161/15384101.2014.988027
Kon S, Ishibashi K, Katoh H et al (2017) Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat Cell Biol 19(5):530–541. https://doi.org/10.1038/ncb3509
Sasaki A, Nagatake T, Egami R et al (2018) Obesity suppresses cell-competition-mediated apical elimination of RasV12-transformed cells from epithelial tissues. Cell Rep 23(4):974–982. https://doi.org/10.1016/j.celrep.2018.03.104
Ayukawa S, Kamoshita N, Nakayama J et al (2021) Epithelial cells remove precancerous cells by cell competition via MHC class I-LILRB3 interaction. Nat Immunol 22(11):1391–1402. https://doi.org/10.1038/s41590-021-01045-6
Takai T (2005) Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology 115(4):433–440. https://doi.org/10.1111/j.1365-2567.2005.02177.x
Yoshihama S, Vijayan S, Sidiq T, Kobayashi KS (2017) NLRC5/CITA: a key player in cancer immune surveillance. Trends in Cancer 3(1):28–38. https://doi.org/10.1016/j.trecan.2016.12.003
Schwartz M, Portugez AS, Attia BZ et al (2020) Genomic retargeting of p53 and CTCF is associated with transcriptional changes during oncogenic HRas-induced transformation. Commun Biol 3 (1). https://doi.org/10.1038/s42003-020-01398-y
Yoh KE, Regunath K, Guzman A et al (2016) Repression of p63 and induction of EMT by mutant Ras in mammary epithelial cells. Proc Natl Acad Sci USA 113(41):E6107–E6116. https://doi.org/10.1073/pnas.1613417113
Franceschi RT, Xiao GZ (2003) Regulation of the osteoblast-specific transcription factor, runx2: responsiveness to multiple signal transduction pathways. J Cell Biochem 88(3):446–454. https://doi.org/10.1002/jcb.10369
Held W, Mariuzza RA (2008) Cis interactions of immunoreceptors with MHC and non-MHC ligands. Nat Rev Immunol 8(4):269–278. https://doi.org/10.1038/nri2278
Shiroishi M, Kuroki K, Rasubala L et al (2006) Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d). Proc Natl Acad Sci USA 103(44):16412–16417. https://doi.org/10.1073/pnas.0605228103
Hill W, Zaragkoulias A, Salvador-Barbero B et al (2021) EPHA2-dependent outcompetition of KRASG12D mutant cells by wild-type neighbors in the adult pancreas. Curr Biol 31(12):2550–2560. https://doi.org/10.1016/j.cub.2021.03.094
Porazinski S, de Navascués J, Yako Y et al (2016) EphA2 drives the segregation of Ras-transformed epithelial cells from normal neighbors. Curr Biol 26(23):3220–3229. https://doi.org/10.1016/j.cub.2016.09.037
Hill W, Hogan C (2019) Normal epithelial cells trigger EphA2-dependent RasV12 cell repulsion at the single cell level. Small GTPases 10(4):305–310. https://doi.org/10.1080/21541248.2017.1324940
Nakamoto M, Cheng HJ, Friedman GC et al (1996) Topographically specific effects of ELF-1 on retinal axon guidance in vitro and retinal axon mapping in vivo. Cell 86(5):755–766. https://doi.org/10.1016/s0092-8674(00)80150-6
Mori Y, Shiratsuchi N, Sato N et al (2022) Extracellular ATP facilitates cell extrusion from epithelial layers mediated by cell competition or apoptosis. Curr Biol 32(10):2144–2159. https://doi.org/10.1016/j.cub.2022.03.057
Ogawa M, Kawarazaki Y, Fujita Y et al (2021) FGF21 induced by the ASK1-p38 pathway promotes mechanical cell competition by attracting cells. Curr Biol 31 (5). https://doi.org/10.1016/j.cub.2020.11.052
Gu YP, Forostyan T, Sabbadini R, Rosenblatt J (2011) Epithelial cell extrusion requires the sphingosine-1-phosphate receptor 2 pathway. J Cell Biol 193(4):667–676. https://doi.org/10.1083/jcb.201010075
Yamamoto S, Yako Y, Fujioka Y et al (2016) A role of the sphingosine-1-phosphate (S1P)-S1P receptor 2 pathway in epithelial defense against cancer (EDAC). Mol Biol Cell 27(3):491–499. https://doi.org/10.1091/mbc.E15-03-0161
Kajita M, Sugimura K, Ohoka A et al (2014) Filamin acts as a key regulator in epithelial defence against transformed cells. Nature Communication 5:4428. https://doi.org/10.1038/ncomms5428
Hamidouche Z, Fromigué O, Ringe J et al (2009) Priming integrin α5 promotes human mesenchymal stromal cell osteoblast differentiation and osteogenesis. Proc Natl Acad Sci USA 106(44):18587–18591. https://doi.org/10.1073/pnas.0812334106
Wu SK, Gomez GA, Michael M et al (2014) Cortical F-actin stabilization generates apical-lateral patterns of junctional contractility that integrate cells into epithelia. Nat Cell Biol 16(2):167–178. https://doi.org/10.1038/ncb2900
Maruyama T, Sasaki A, Iijima S et al (2020) ZAK inhibitor PLX4720 promotes extrusion of transformed cells via cell competition. iScience 23 (7). https://doi.org/10.1016/j.isci.2020.101327
Sato N, Yako Y, Maruyama T et al (2020) The COX-2/PGE2 pathway suppresses apical elimination of RasV12-transformed cells from epithelia. Commun Biol 3 (1). https://doi.org/10.1038/s42003-020-0847-y
Gallini S, Annusver K, Rahman NT et al (2023) Injury prevents Ras mutant cell expansion in mosaic skin. Nature 619:167–175. https://doi.org/10.1038/s41586-023-06198-y
Greten FR, Grivennikov SI (2019) Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51(1):27–41. https://doi.org/10.1016/j.immuni.2019.06.025
Pothapragada SP, Gupta P, Mukherjee S and Das T (2022) Matrix mechanics regulates epithelial defence against cancer by tuning dynamic localization of filamin. Nat Commun 13 (1). https://doi.org/10.1038/s41467-021-27896-z
Madan E, Pelham CJ, Nagane M et al (2019) Flower isoforms promote competitive growth in cancer. Nature 572(7768):260–264. https://doi.org/10.1038/s41586-019-1429-3
Rhiner C, López-Gay JM, Soldini D et al (2010) Flower forms an extracellular code that reveals the fitness of a cell to its neighbors in Drosophila. Dev Cell 18(6):985–998. https://doi.org/10.1016/j.devcel.2010.05.010
de la Cova C, Abril M, Bellosta P et al (2004) Drosophila Myc regulates organ size by inducing cell competition. Cell 117(1):107–116. https://doi.org/10.1016/s0092-8674(04)00214-4
Moreno E, Basler K (2004) dMyc transforms cells into super-competitors. Cell 117(1):117–129. https://doi.org/10.1016/s0092-8674(04)00262-4
Ziosi M, Baena-López LA, Grifoni D et al (2010) dMyc functions downstream of Yorkie to promote the supercompetitive behavior of Hippo pathway mutant cells. PLoS Genet 6 (9). https://doi.org/10.1371/journal.pgen.1001140
Rodrigues AB, Zoranovic T, Ayala-Camargo A et al (2012) Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie. Wingless and ribosome biogenesis Development 139(21):4051–4061. https://doi.org/10.1242/dev.076760
Di Giacomo S, Sollazzo M, de Biase D et al (2017) Human cancer cells signal their competitive fitness through MYC activity. Sci Rep 7. https://doi.org/10.1038/s41598-017-13002-1
Patel MS, Shah HS, Shrivastava N (2017) c-Myc-dependent cell competition in human cancer cells. J Cell Biochem 118(7):1782–1791. https://doi.org/10.1002/jcb.25846
Liu ZJ, Yee PP, Wei YJ et al (2019) Differential YAP expression in glioma cells induces cell competition and promotes tumorigenesis. J Cell Sci 132 (5). https://doi.org/10.1242/jcs.225714.
Flanagan DJ, Pentinmikko N, Luopajärvi K et al (2021) NOTUM from Apc-mutant cells biases clonal competition to initiate cancer. Nature 594(7863):430–435. https://doi.org/10.1038/s41586-021-03525-z
van Neerven SM, de Groot NE, Nijman LE et al (2021) Apc-mutant cells act as supercompetitors in intestinal tumour initiation. Nature 594(7863):436–441. https://doi.org/10.1038/s41586-021-03558-4
Kohashi K, Mori Y, Narumi R et al (2021) Sequential oncogenic mutations influence cell competition. Curr Biol 31(18):3984–3995. https://doi.org/10.1016/j.cub.2021.06.064
Kullander K, Klein R (2002) Mechanisms and functions of EPH and ephrin signalling. Nat Rev Mol Cell Biol 3(7):475–486. https://doi.org/10.1038/nrm856
Geng LL, Lam KSL, Xu AM (2020) The therapeutic potential of FGF21 in metabolic diseases: from bench to clinic. Nat Rev Endocrinol 16(11):654–667. https://doi.org/10.1038/s41574-020-0386-0
Jhunjhunwala S, Hammer C, Delamarre L (2021) Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer 21(5):298–312. https://doi.org/10.1038/s41568-021-00339-z
Geng J, Raghavan M (2021) Conformational sensing of major histocompatibility complex (MHC) class I molecules by immune receptors and intracellular assembly factors. Curr Opin Immunol 70:67–74. https://doi.org/10.1016/j.coi.2021.03.014
Aptsiauri NT, Cabrera A, Garcia-Lora MA et al (2007) MHC class I antigens and immune surveillance in transformed cells. Int Rev Cytol 256:139–189. https://doi.org/10.1016/s0074-7696(07)56005-5
Ljunggren HG, Karre K (1990) In search of the missing self — MHC molecules and NK cell recognition. Immunol Today 11(7):237–244. https://doi.org/10.1016/0167-5699(90)90097-s
Norris JL, Baldwin AS (1999) Oncogenic Ras enhances NF-κB transcriptional activity through Raf-dependent and Raf-independent mitogen-activated protein kinase signaling pathways. J Biol Chem 274(20):13841–13846. https://doi.org/10.1074/jbc.274.20.13841
Artis D (2008) Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol 8(6):411–420. https://doi.org/10.1038/nri2316
Bastounis EE, Serrano-Alcalde F, Radhakrishnan P et al (2021) Mechanical competition triggered by innate immune signaling drives the collective extrusion of bacterially infected epithelial cells. Dev Cell 56(4):443–460. https://doi.org/10.1016/j.devcel.2021.01.012
Gobin SJP, Keijsers V, van Zutphen M et al (1998) The role of enhancer A in the locus-specific transactivation of classical and nonclassical HLA class I genes by nuclear factor κB. J Immunol 161(5):2276–2283. https://doi.org/10.4049/jimmunol.161.5.2276
Guo XC, Liu TX, Shi HF et al (2015) Respiratory syncytial virus infection upregulates NLRC5 and major histocompatibility complex class I expression through RIG-I induction in airway epithelial cells. J Virol 89(15):7636–7645. https://doi.org/10.1128/jvi.00349-15
Herzer K, Falk CS, Encke J et al (2003) Upregulation of major histocompatibility complex class I on liver cells by hepatitis C virus core protein via p53 and TAP1 impairs natural killer cell cytotoxicity. J Virol 77(15):8299–8309. https://doi.org/10.1128/jvi.77.15.8299-8309.2003
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (B) (Grant Number 18H02675), the Fusion Oriented REsearch for disruptive Science and Technology (FOREST) (Grant Number JPMJFR226D) from the Japan Science and Technology Agency (JST), Advanced Research & Development Programs for Medical Innovation (Prime) (Grant Number 19gm6210019h0001), and Project for Promotion of Cancer Research and Therapeutic Evolution (P-PROMOTE) (Grant Number 23ama221113h0002) from the Japan Agency for Medical Research and Development (AMED), Takeda Science Foundation, the Naito Foundation, and the TERUMO Life Science foundation (to T. M.).
Author information
Authors and Affiliations
Contributions
Conceptualisation: T. M.; writing: S. A., N. K., and T. M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is a contribution to the Article Collection on Immunopathology of Barrier Function – Guest Editor: Koji Hase & Hiroshi Ohno
The original online version of this article was revised: In this article the authors’ proof corrections were incorrectly carried out. Among some minor changes to the text, the following major revisions took place:
Affiliation “Waseda Institute for Advanced Study, Waseda University, Tokyo, Japan” was added to authors Nagisa Kamoshita and Takeshi Maruyama.
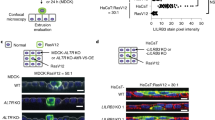
Figure 1A: The dashed vertical axis was hidden in the upper part of Fig. 1A.
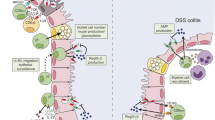
Figure 2C: The vertical axis was hidden in the right hand graphic of Fig. 2C.
Figure 3B: “ephrin-A” was incorrectly written as “ephrinA”.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ayukawa, S., Kamoshita, N. & Maruyama, T. Epithelial recognition and elimination against aberrant cells. Semin Immunopathol (2024). https://doi.org/10.1007/s00281-024-01001-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00281-024-01001-0