Abstract
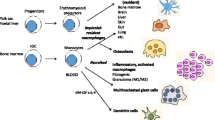
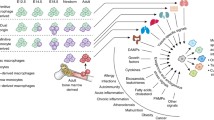
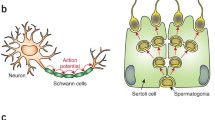
The vast majority of macrophages in the body are present in tissues outside the circulation, yet their remarkable phenotypic heterogeneity within different anatomical compartments in the steady state and disease remains essentially unexplored. The development of immunocytochemical and fluorescent histological methods has revealed the presence and morphological diversity of macrophages within tissues, especially in the mouse, but functional specialisation in situ is difficult to establish and their mechanistic basis remains unknown, particularly in humans, in spite of the rich material available. In this review, we consider some of the technical issues that hamper investigation, assess the contribution of cell differentiation and modulation to macrophage heterogeneity within tissues and illustrate variation of macrophages and closely related cells in selected organs in mouse and man.



Similar content being viewed by others
References
Ginhoux F et al (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330(6005):841–845
Merad M et al (2002) Langerhans cells renew in the skin throughout life under steady-state conditions. Nat Immunol 3(12):1135–1141
Schulz C et al (2012) A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336(6077):86–90
Yona S et al (2013) Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38(1):79–91
Swirski FK et al (2009) Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 325(5940):612–616
Geissmann F et al (2010) Development of monocytes, macrophages, and dendritic cells. Science 327(5966):656–661
Jenkins SJ et al (2011) Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332(6035):1284–1288
Meredith MM et al (2012) Zinc finger transcription factor zDC is a negative regulator required to prevent activation of classical dendritic cells in the steady state. J Exp Med 209(9):1583–1593
Satpathy AT et al (2012) Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med 209(6):1135–1152
Kaplan G (2013) Macrophages in tuberculosis: friend or foe. Semin Immunopathol. This Issue
Libby P (2013) Monocyte heterogeneity in cardiovascular disease. Semin Immunopathol. This Issue
Biswas SK, Allavena P, Mantovani A (2013) Tumour-associated macrophages: functional diversity, clinical significance and open questions. Semin Immunopathol. doi:10.1007/s00281-013-0367-7
den Haan JMM, Martinez-Pomares L (2013) Macrophage heterogeneity in lymphoid tissues. Semin Immunopathol. doi:10.1007/s00281-013-0378-4
Perry VH (2013) Microglia in chronic neurodegeneration: the contribution of microglia priming and systemic inflammation. Semin Immunopathol. doi:10.1007/s00281-013-0382-8
Gordon S et al (2011) F4/80 and the related adhesion-GPCRs. Eur J Immunol 41(9):2472–2476
Austyn JM, Gordon S (1981) F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol 11(10):805–815
Hume DA (2013) The macrophage. http://www.macrophages.com/home. Accessed 9 May
Lin HH et al (2005) The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J Exp Med 201(10):1615–1625
Crocker PR, Gordon S (1989) Mouse macrophage hemagglutinin (sheep erythrocyte receptor) with specificity for sialylated glycoconjugates characterized by a monoclonal antibody. J Exp Med 169(4):1333–1346
Taylor PR et al (2005) Macrophage receptors and immune recognition. Annu Rev Immunol 23:901–944
van der Laan LJ et al (1997) Macrophage scavenger receptor MARCO: in vitro and in vivo regulation and involvement in the anti-bacterial host defense. Immunol Lett 57(1–3):203–208
Keshav S et al (1991) Lysozyme is an inducible marker of macrophage activation in murine tissues as demonstrated by in situ hybridization. J Exp Med 174(5):1049–1058
Geissmann F, Jung S, Littman DR (2003) Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19(1):71–82
Egen JG et al (2008) Macrophage and T cell dynamics during the development and disintegration of mycobacterial granulomas. Immunity 28(2):271–284
Collin M et al (2013) Monocytes, macrophages, and dendritic cells. In: Wintrobe’s clinical hematology. Lippincott Williams & Wilkins, Philadelphia
Chavele KM et al (2010) Mannose receptor interacts with Fc receptors and is critical for the development of crescentic glomerulonephritis in mice. J Clin Invest 120(5):1469–1478
Cohen NR et al (2013) Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol 14(1):90–99
Miller JC et al (2012) Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol 13(9):888–899
Martinez FO et al (2013) Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood 121(9):e57–e69
Kohyama M et al (2009) Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature 457(7227):318–321
Perez OD, Nolan GP (2006) Phospho-proteomic immune analysis by flow cytometry: from mechanism to translational medicine at the single-cell level. Immunol Rev 210:208–228
Panicker LM et al (2012) Induced pluripotent stem cell model recapitulates pathologic hallmarks of Gaucher disease. Proc Natl Acad Sci U S A
Gerner MY et al (2012) Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37(2):364–376
Acknowledgments
The preparation of the manuscript was undertaken by SG while a fellow of the Stellenbosch Institute of Advanced Study.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is a contribution to the special issue on Macrophage Heterogeneity, Subsets and Human Disease - Guest Editor: Siamon Gordon
Rights and permissions
About this article
Cite this article
Gordon, S., Plűddemann, A. Tissue macrophage heterogeneity: issues and prospects. Semin Immunopathol 35, 533–540 (2013). https://doi.org/10.1007/s00281-013-0386-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-013-0386-4