Abstract
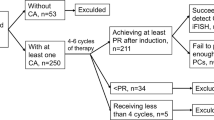
We analyzed variations in terms of chromosomal abnormalities (CA) by fluorescence in situ hybridization (FISH) analysis on purified bone marrow plasma cells throughout the progression from monoclonal gammopathy of undetermined significance/smoldering multiple myeloma (MGUS/SMM) to newly diagnosed MM/plasma cell leukemia (NDMM/PCL) at diagnosis and from diagnostic samples to progressive disease. High risk was defined by the presence of at least del(17p), t(4;14), and/or t(14;16). 1p/1q detection (in the standard FISH panel from 2012 onward) was not available for all patients. We analyzed 139 MM/PCL diagnostic samples from 144 patients, with a median follow-up of 71 months: high-risk CA at diagnosis (MGUS/SMM or NDMM) was present in 28% of samples, whereas 37–39% showed high-risk CA at relapse. In 115 patients with NDMM who evolved to relapsed/refractory MM, we identified 3 different populations: (1) 31/115 patients (27%) with gain of new CA (del13, del17p, t(4;14), t(14;16) or 1q CA when available); (2) 10/115 (9%) patients with loss of a previously identified CA; and (3) 74 patients with no changes. The CA gain group showed a median overall survival of 66 months vs. 84 months in the third group (HR 0.56, 95% CI 0.34–0.92, p = 0.023). Clonal evolution occurs as disease progresses after different chemotherapy lines. Patients who acquired high-risk CA had the poorest prognosis. Our findings highlight the importance of performing FISH analysis both at diagnosis and at relapse.


Similar content being viewed by others
Data availability
After the publication of this article, data collected for this retrospective analysis and related documents will be made available to others upon reasonably justified request, which has to be written and addressed to the attention of the corresponding author Dr. Stefania Oliva at the following e-mail address: stefania.olivamolinet@gmail.com. The corresponding author Dr. Stefania Oliva is responsible to evaluate and eventually accept or refuse every request to disclose data and their related documents, in compliance with the ethical approval conditions, in compliance with applicable laws and regulations, and in conformance with the agreements in place with the involved subjects, the participating institutions, and all the other parties directly or indirectly involved in the participation, conduct, development, management, and evaluation of this analysis.
References
Bolli N, Avet-Loiseau H, Wedge DC, van Loo P, Alexandrov LB, Martincorena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, Hinton JW, Li Y, Tubio JMC, McLaren S, O' Meara S, Butler AP, Teague JW, Mudie L, Anderson E, Rashid N, Tai YT, Shammas MA, Sperling AS, Fulciniti M, Richardson PG, Parmigiani G, Magrangeas F, Minvielle S, Moreau P, Attal M, Facon T, Futreal PA, Anderson KC, Campbell PJ, Munshi NC (2014) Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 5:2997. https://doi.org/10.1038/ncomms3997
Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, Cibulskis K, McKenna A, Chapman MA, Straussman R, Levy J, Perkins LM, Keats JJ, Schumacher SE, Rosenberg M, Getz G, Golub TR, Anderson KC, Richardson P, Krishnan A, Lonial S, Kaufman J, Siegel DS, Vesole DH, Roy V, Rivera CE, Rajkumar SV, Kumar S, Fonseca R, Ahmann GJ, Bergsagel PL, Stewart AK, Hofmeister CC, Efebera YA, Jagannath S, Chari A, Trudel S, Reece D, Wolf J, Martin T, Zimmerman T, Rosenbaum C, Jakubowiak AJ, Lebovic D, Vij R, Stockerl-Goldstein K (2014) Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 25:91–101. https://doi.org/10.1016/J.CCR.2013.12.015
Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, Proszek PZ, Johnson DC, Kaiser MF, Melchor L, Aronson LI, Scales M, Pawlyn C, Mirabella F, Jones JR, Brioli A, Mikulasova A, Cairns DA, Gregory WM, Quartilho A, Drayson MT, Russell N, Cook G, Jackson GH, Leleu X, Davies FE, Morgan GJ (2015) Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol 33:3911–3920. https://doi.org/10.1200/JCO.2014.59.1503
Fonseca R (2003) Many and multiple myeloma(s). Leukemia 17:1943–1944. https://doi.org/10.1038/sj.leu.2403090
Fonseca R, Barlogie B, Bataille R et al (2004) Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res 64:1546–1558. https://doi.org/10.1158/0008-5472.can-03-2876
Morgan GJ, Walker BA, Davies FE (2012) The genetic architecture of multiple myeloma. Nat Rev Cancer 12:335–348. https://doi.org/10.1038/nrc3257
Nowell PC (1976) The clonal evolution of tumor cell populations. Science (80- ) 194:23–28. https://doi.org/10.1126/science.959840
Burrell RA, McGranahan N, Bartek J, Swanton C (2013) The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501:338–345. https://doi.org/10.1038/nature12625
Corre J, Munshi N, Avet-Loiseau H (2015) Genetics of multiple myeloma: another heterogeneity level? Blood 125:1870–1876. https://doi.org/10.1182/blood-2014-10-567370
Robiou Du Pont S, Cleynen A, Fontan C et al (2017) Genomics of multiple myeloma. J Clin Oncol 35:963–967. https://doi.org/10.1200/jco.2016.70.6705
Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, Morgan G, van Ness B, Chesi M, Minvielle S, Neri A, Barlogie B, Kuehl WM, Liebisch P, Davies F, Chen-Kiang S, Durie BG, Carrasco R, Sezer O, Reiman T, Pilarski L, Avet-Loiseau H, International Myeloma Working Group (2009) International myeloma working group molecular classification of multiple myeloma: spotlight review. Leukemia 23:2210–2221. https://doi.org/10.1038/leu.2009.174
Mikhael JR, Dingli D, Roy V, Reeder CB, Buadi FK, Hayman SR, Dispenzieri A, Fonseca R, Sher T, Kyle RA, Lin Y, Russell SJ, Kumar S, Bergsagel PL, Zeldenrust SR, Leung N, Drake MT, Kapoor P, Ansell SM, Witzig TE, Lust JA, Dalton RJ, Gertz MA, Stewart AK, Rajkumar SV, Chanan-Khan A, Lacy MQ, Mayo Clinic (2013) Management of newly diagnosed symptomatic multiple myeloma: updated Mayo stratification of myeloma and risk-adapted therapy (mSMART) consensus guidelines 2013. Mayo Clin Proc 88:360–376. https://doi.org/10.1016/j.mayocp.2013.01.019
Avet-Loiseau H, Attal M, Campion L, Caillot D, Hulin C, Marit G, Stoppa AM, Voillat L, Wetterwald M, Pegourie B, Voog E, Tiab M, Banos A, Jaubert J, Bouscary D, Macro M, Kolb B, Traulle C, Mathiot C, Magrangeas F, Minvielle S, Facon T, Moreau P (2012) Long-term analysis of the ifm 99 trials for myeloma: cytogenetic abnormalities [t(4;14), del(17p), 1q gains] play a major role in defining long-term survival. J Clin Oncol 30:1949–1952. https://doi.org/10.1200/JCO.2011.36.5726
Zandecki M, Laï JL, Geneviève F et al (1997) Several cytogenetic subclones may be identified within plasma cells from patients with monoclonal gammopathy of undetermined significance, both at diagnosis and during the indolent course of this condition. Blood 90:3682–3690
López-Corral L, Gutiérrez NC, Vidriales MB et al (2011) The progression from MGUS to smoldering myeloma and eventually to multiple myeloma involves a clonal expansion of genetically abnormal plasma cells. Clin Cancer Res 17:1692–1700. https://doi.org/10.1158/1078-0432.CCR-10-1066
Drach J, Angerler J, Schuster J, Rothermundt C, Thalhammer R, Haas OA, Jäger U, Fiegl M, Geissler K, Ludwig H, Huber H (1995) Interphase fluorescence in situ hybridization identifies chromosomal abnormalities in plasma cells from patients with monoclonal gammopathy of undetermined significance. Blood 86:3915–3921
Zhao S, Choi M, Heuck C, Mane S, Barlogie B, Lifton RP, Dhodapkar MV (2014) Serial exome analysis of disease progression in premalignant gammopathies. Leukemia 28:1548–1552. https://doi.org/10.1038/leu.2014.59
Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, Mirabella F, Lopez-Corral L, Humphray S, Murray L, Ross M, Bentley D, Gutiérrez NC, Garcia-Sanz R, San Miguel J, Davies FE, Gonzalez D, Morgan GJ (2014) Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia 28:384–390. https://doi.org/10.1038/leu.2013.199
Mikulasova A, Wardell CP, Murison A, Boyle EM, Jackson GH, Smetana J, Kufova Z, Pour L, Sandecka V, Almasi M, Vsianska P, Gregora E, Kuglik P, Hajek R, Davies FE, Morgan GJ, Walker BA (2017) The spectrum of somatic mutations in monoclonal gammopathy of undetermined significance indicates a less complex genomic landscape than that in multiple myeloma. Haematologica 102:1617–1625. https://doi.org/10.3324/haematol.2017.163766
Dutta AK, Fink JL, Grady JP, Morgan GJ, Mullighan CG, To LB, Hewett DR, Zannettino ACW (2019) Subclonal evolution in disease progression from MGUS/SMM to multiple myeloma is characterised by clonal stability. Leukemia 33:457–468. https://doi.org/10.1038/s41375-018-0206-x
Bolli N, Maura F, Minvielle S, Gloznik D, Szalat R, Fullam A, Martincorena I, Dawson KJ, Samur MK, Zamora J, Tarpey P, Davies H, Fulciniti M, Shammas MA, Tai YT, Magrangeas F, Moreau P, Corradini P, Anderson K, Alexandrov L, Wedge DC, Avet-Loiseau H, Campbell P, Munshi N (2018) Genomic patterns of progression in smoldering multiple myeloma. Nat Commun 9:3363. https://doi.org/10.1038/s41467-018-05058-y
Ledergor G, Weiner A, Zada M, Wang SY, Cohen YC, Gatt ME, Snir N, Magen H, Koren-Michowitz M, Herzog-Tzarfati K, Keren-Shaul H, Bornstein C, Rotkopf R, Yofe I, David E, Yellapantula V, Kay S, Salai M, Ben Yehuda D, Nagler A, Shvidel L, Orr-Urtreger A, Halpern KB, Itzkovitz S, Landgren O, San-Miguel J, Paiva B, Keats JJ, Papaemmanuil E, Avivi I, Barbash GI, Tanay A, Amit I (2018) Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med 24:1867–1876. https://doi.org/10.1038/s41591-018-0269-2
Mailankody S, Kazandjian D, Korde N, Roschewski M, Manasanch E, Bhutani M, Tageja N, Kwok M, Zhang Y, Zingone A, Lamy L, Costello R, Morrison C, Hultcrantz M, Christofferson A, Washington M, Boateng M, Steinberg SM, Stetler-Stevenson M, Figg WD, Papaemmanuil E, Wilson WH, Keats JJ, Landgren O (2017) Baseline mutational patterns and sustained MRD negativity in patients with high-risk smoldering myeloma. Blood Adv 1:1911–1918. https://doi.org/10.1182/bloodadvances.2017005934
Gerber B, Manzoni M, Spina V, Bruscaggin A, Lionetti M, Fabris S, Barbieri M, Ciceri G, Pompa A, Forestieri G, Lerch E, Servida P, Bertoni F, Zucca E, Ghielmini M, Cortelezzi A, Cavalli F, Stussi G, Baldini L, Rossi D, Neri A (2018) Circulating tumor DNA as a liquid biopsy in plasma cell dyscrasias. Haematologica 103:e245–e248. https://doi.org/10.3324/haematol.2017.184358
Corre J, Cleynen A, Robiou du Pont S, Buisson L, Bolli N, Attal M, Munshi N, Avet-Loiseau H (2018) Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia 32:2636–2647. https://doi.org/10.1038/s41375-018-0153-6
Bergsagel PL, Zhan F, Sawyer J et al (2005) Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 106:296–303. https://doi.org/10.1182/blood-2005-01-0034
Hébraud B, Caillot D, Corre J et al (2013) The translocation t(4;14) can be present only in minor subclones in multiple myeloma. Clin Cancer Res 19:4634–4637. https://doi.org/10.1158/1078-0432.CCR-12-3866
Acknowledgments
We thank all the patients involved in this analysis, the nurses Manuela Grasso and Luca Merlone, the statistician Stefano Spada, the data manager Marco Poggiu, and Ugo Panzani from the Torino site.
Author information
Authors and Affiliations
Contributions
SO and PO conceived and designed the work that led to the submission.
All the authors acquired the data and interpreted the results.
SO drafted the first version of the manuscript.
All the authors revised the manuscript and approved the final version.
All the authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of interest
SO has received honoraria from Amgen, Celgene, and Janssen and has served on the advisory boards for Adaptive Biotechnologies, Janssen, Amgen, and Takeda. MD has received honoraria for lectures from Sanofi and GSK and has served on the advisory boards for GSK. RM has received honoraria from Sanofi, Celgene, Takeda, and Janssen; has served on the advisory boards for Sanofi, Takeda, and Janssen; and has received consultancy fees from Janssen. SB has received honoraria from Celgene, Amgen, Janssen, and Bristol-Myers Squibb; has served on the advisory boards for Celgene, Amgen, Janssen, and Karyopharm; and has received consultancy fees from Janssen and Takeda. MB has received honoraria from Sanofi, Celgene, Amgen, Janssen, Novartis, Bristol-Myers Squibb, and AbbVie; has served on the advisory boards for Janssen and GSK; and has received research funding from Sanofi, Celgene, Amgen, Janssen, Novartis, Bristol-Myers Squibb, and Mundipharma. PO has served on the advisory boards for Janssen. The other authors declare no competing financial interests.
Ethics approval
All procedures followed were in accordance with the ethical standards of the responsible committees on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008. Patients gave written informed consent before bone marrow aspiration and before starting therapy.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Oliva, S., De Paoli, L., Ruggeri, M. et al. A longitudinal analysis of chromosomal abnormalities in disease progression from MGUS/SMM to newly diagnosed and relapsed multiple myeloma. Ann Hematol 100, 437–443 (2021). https://doi.org/10.1007/s00277-020-04384-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-020-04384-w