Abstract
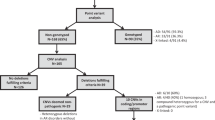
BS69 was originally identified as an adenovirus E1A-binding protein and was found to be involved in multiple cellular events. A recent array-based study implicated the presence of copy number variations (CNVs) of BS69 in the genomes of acute myelogenous leukemia. CNVs are present in the general population at varying degrees and have been found to associate with various types of diseases including hematological malignancies. However, most of the current studies focused on the genome-wide screening of CNVs, and the functional impact of such regions needs to be extensively investigated in large amount of clinical samples. Thus, in our study, we collected 617 bone marrow samples from multi-types of hematological malignancies as well as healthy controls. We found significant association between the CNVs of BS69 and these hematological malignancies including acute lymphoblastic leukemia (ALL), acute myelogenous leukemia (AML), chronic lymphocytic leukemia (CLL), chronic myelogenous leukemia (CML), multiple myeloma (MM), and myelodysplastic syndrome (MDS). We also examined the expression of BS69 mRNA in the samples with one or two copies of DNA, and observed a weak yet positive correlation between the relative expression level and gene dosage. In general, the CNVs of BS69 have the potential to serve as a diagnostic indicator, alone or in combination with other markers, for hematological malignancies.

Similar content being viewed by others

References
Hateboer G, Gennissen A, Ramos YF, Kerkhoven RM, Sonntag-Buck V, Stunnenberg HG, Bernards R (1995) BS69, a novel adenovirus E1A-associated protein that inhibits E1A transactivation. EMBO J 14:3159–3169
Ikeda O, Miyasaka Y, Yoshida R, Mizushima A, Oritani K, Sekine Y, Kuroda M, Yasui T, Fujimuro M, Muromoto R, Nanbo A, Matsuda T: BS69 cooperates with TRAF3 in the regulation of Epstein-Barr virus-derived LMP1/CTAR1-induced NF-kappaB activation. FEBS Lett
Ikeda O, Sekine Y, Mizushima A, Oritani K, Yasui T, Fujimuro M, Muromoto R, Nanbo A, Matsuda T (2009) BS69 negatively regulates the canonical NF-kappaB activation induced by Epstein-Barr virus-derived LMP1. FEBS Lett 583:1567–1574
Wan J, Zhang W, Wu L, Bai T, Zhang M, Lo KW, Chui YL, Cui Y, Tao Q, Yamamoto M, Akira S, Wu Z (2006) BS69, a specific adaptor in the latent membrane protein 1-mediated c-Jun N-terminal kinase pathway. Mol Cell Biol 26:448–456
Masselink H, Bernards R (2000) The adenovirus E1A binding protein BS69 is a corepressor of transcription through recruitment of N-CoR. Oncogene 19:1538–1546
Ekblad CM, Chavali GB, Basu BP, Freund SM, Veprintsev D, Hughes-Davies L, Kouzarides T, Doherty AJ, Itzhaki LS (2005) Binding of EMSY to HP1beta: implications for recruitment of HP1beta and BS69. EMBO Rep 6:675–680
Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, Nielsen T, Schulzer M, Chia S, Ragaz J, Cahn A, Linger L, Ozdag H, Cattaneo E, Jordanova ES, Schuuring E, Yu DS, Venkitaraman A, Ponder B, Doherty A, Aparicio S, Bentley D, Theillet C, Ponting CP, Caldas C, Kouzarides T (2003) EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell 115:523–535
Ladendorff NE, Wu S, Lipsick JS (2001) BS69, an adenovirus E1A-associated protein, inhibits the transcriptional activity of c-Myb. Oncogene 20:125–132
Masselink H, Vastenhouw N, Bernards R (2001) B-myb rescues ras-induced premature senescence, which requires its transactivation domain. Cancer Lett 171:87–101
Velasco G, Grkovic S, Ansieau S (2006) New insights into BS69 functions. J Biol Chem 281:16546–16550
Wei G, Schaffner AE, Baker KM, Mansky KC, Ostrowski MC (2003) Ets-2 interacts with co-repressor BS69 to repress target gene expression. Anticancer Res 23:2173–2178
Zhang W, Chan HM, Gao Y, Poon R, Wu Z (2007) BS69 is involved in cellular senescence through the p53-p21Cip1 pathway. EMBO Rep 8:952–958
Yu B, Shao Y, Zhang C, Chen Y, Zhong Q, Zhang J, Yang H, Zhang W, Wan J (2009) BS69 undergoes SUMO modification and plays an inhibitory role in muscle and neuronal differentiation. Exp Cell Res 315:3543–3553
Ulger C, Toruner GA, Alkan M, Mohammed M, Damani S, Kang J, Galante A, Aviv H, Soteropoulos P, Tolias PP, Schwalb MN, Dermody JJ (2003) Comprehensive genome-wide comparison of DNA and RNA level scan using microarray technology for identification of candidate cancer-related genes in the HL-60 cell line. Cancer Genet Cytogenet 147:28–35
Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, Baty J, Heath S, Westervelt P, Watson MA, Tomasson MH, Nagarajan R, O'Gara BP, Bloomfield CD, Mrozek K, Selzer RR, Richmond TA, Kitzman J, Geoghegan J, Eis PS, Maupin R, Fulton RS, McLellan M, Wilson RK, Mardis ER, Link DC, Graubert TA, DiPersio JF, Ley TJ (2009) Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA 106:12950–12955
Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK (2006) A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 38:75–81
Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA (2006) Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet 38:82–85
Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951
Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE (2008) Mapping and sequencing of structural variation from eight human genomes. Nature 453:56–64
Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, Taillon BE, Chen Z, Tanzer A, Saunders AC, Chi J, Yang F, Carter NP, Hurles ME, Weissman SM, Harkins TT, Gerstein MB, Egholm M, Snyder M (2007) Paired-end mapping reveals extensive structural variation in the human genome. Science 318:420–426
McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, Altshuler DM (2006) Common deletion polymorphisms in the human genome. Nat Genet 38:86–92
Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME (2006) Global variation in copy number in the human genome. Nature 444:444–454
Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528
Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE (2005) Fine-scale structural variation of the human genome. Nat Genet 37:727–732
Korbel JO, Urban AE, Grubert F, Du J, Royce TE, Starr P, Zhong G, Emanuel BS, Weissman SM, Snyder M, Gerstein MB (2007) Systematic prediction and validation of breakpoints associated with copy-number variants in the human genome. Proc Natl Acad Sci USA 104:10110–10115
Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA (2005) O'Connell R J, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK: The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307:1434–1440
McKinney C, Merriman ME, Chapman PT, Gow PJ, Harrison AA, Highton J, Jones PB, McLean L, O'Donnell JL, Pokorny V, Spellerberg M, Stamp LK, Willis J, Steer S, Merriman TR (2008) Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis 67:409–413
Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D (2006) APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 38:24–26
Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K (2003) Alpha-Synuclein locus triplication causes Parkinson’s disease. Science 302:841
Andrews J, Kennette W, Pilon J, Hodgson A, Tuck AB, Chambers AF, Rodenhiser DI Multi-platform whole-genome microarray analyses refine the epigenetic signature of breast cancer metastasis with gene expression and copy number. PLoS One 5:e8665
Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, Cole K, Mosse YP, Wood A, Lynch JE, Pecor K, Diamond M, Winter C, Wang K, Kim C, Geiger EA, McGrady PW, Blakemore AI, London WB, Shaikh TH, Bradfield J, Grant SF, Li H, Devoto M, Rappaport ER, Hakonarson H, Maris JM (2009) Copy number variation at 1q21.1 associated with neuroblastoma. Nature 459:987–991
Liu W, Sun J, Li G, Zhu Y, Zhang S, Kim ST, Sun J, Wiklund F, Wiley K, Isaacs SD, Stattin P, Xu J, Duggan D, Carpten JD, Isaacs WB, Gronberg H, Zheng SL, Chang BL (2009) Association of a germ-line copy number variation at 2p24.3 and risk for aggressive prostate cancer. Cancer Res 69:2176–2179
Grubor V, Krasnitz A, Troge JE, Meth JL, Lakshmi B, Kendall JT, Yamrom B, Alex G, Pai D, Navin N, Hufnagel LA, Lee YH, Cook K, Allen SL, Rai KR, Damle RN, Calissano C, Chiorazzi N, Wigler M, Esposito D (2009) Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA). Blood 113:1294–1303
Gunnarsson R, Staaf J, Jansson M, Ottesen AM, Goransson H, Liljedahl U, Ralfkiaer U, Mansouri M, Buhl AM, Smedby KE, Hjalgrim H, Syvanen AC, Borg A, Isaksson A, Jurlander J, Juliusson G, Rosenquist R (2008) Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia–a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer 47:697–711
Schafer M, Schwender H, Merk S, Haferlach C, Ickstadt K, Dugas M (2009) Integrated analysis of copy number alterations and gene expression: a bivariate assessment of equally directed abnormalities. Bioinformatics 25:3228–3235
Strefford JC, van Delft FW, Robinson HM, Worley H, Yiannikouris O, Selzer R, Richmond T, Hann I, Bellotti T, Raghavan M, Young BD, Saha V, Harrison CJ (2006) Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc Natl Acad Sci USA 103:8167–8172
Sulong S, Moorman AV, Irving JA, Strefford JC, Konn ZJ, Case MC, Minto L, Barber KE, Parker H, Wright SL, Stewart AR, Bailey S, Bown NP, Hall AG, Harrison CJ (2009) A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood 113:100–107
Dear PH (2009) Copy-number variation: the end of the human genome? Trends Biotechnol 27:448–454
Shlien A, Tabori U, Marshall CR, Pienkowska M, Feuk L, Novokmet A, Nanda S, Druker H, Scherer SW, Malkin D (2008) Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci USA 105:11264–11269
Dermitzakis ET, Stranger BE (2006) Genetic variation in human gene expression. Mamm Genome 17:503–508
Reymond A, Henrichsen CN, Harewood L, Merla G (2007) Side effects of genome structural changes. Curr Opin Genet Dev 17:381–386
Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SA, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, Hubner N, Cuppen E (2008) Distribution and functional impact of DNA copy number variation in the rat. Nat Genet 40:538–545
Henrichsen CN, Vinckenbosch N, Zollner S, Chaignat E, Pradervand S, Schutz F, Ruedi M, Kaessmann H, Reymond A (2009) Segmental copy number variation shapes tissue transcriptomes. Nat Genet 41:424–429
Sexton T, Umlauf D, Kurukuti S, Fraser P (2007) The role of transcription factories in large-scale structure and dynamics of interphase chromatin. Semin Cell Dev Biol 18:691–697
Author information
Authors and Affiliations
Corresponding authors
Additional information
Hong Yang and Chao Zhang contributed equally to this work.
Jun Wan and Xiaojun Huang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yang, H., Zhang, C., Zhao, X. et al. Analysis of copy number variations of BS69 in multiple types of hematological malignancies. Ann Hematol 89, 959–964 (2010). https://doi.org/10.1007/s00277-010-0966-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-010-0966-5