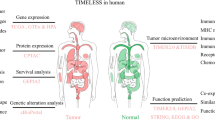
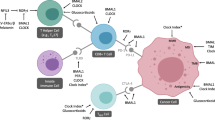
Abstract
Background
While epidemiological studies have established a firm link between circadian disruption and tumorigenesis, the role and mechanism are not fully understood, complicating the design of therapeutic targets related to circadian rhythms (CR). Here, we aimed to explore the intertumoral heterogeneity of CR and elucidate its impact on the tumor microenvironment (TME), drug sensitivity, and immunotherapy.
Methods
Based on unsupervised clustering of 28 CR genes, two distinct CR subtypes (cluster-A and cluster-B) were identified in the TCGA cohort. We further constructed a circadian rhythm signature (CRS) based on the CR genes primarily responsible for clustering to quantify CR activity and to distinguish CR subtypes of individual patients from external datasets. CR subtypes were evaluated by TME characteristics, functional annotation, clinical features, and therapeutic response.
Results
The cluster-B (low-CRS) group was characterized by highly enriched immune-related pathways, high immune cell infiltration, and high anti-tumor immunity, while the cluster-A (high-CRS) group was associated with immunosuppression, synaptic transmission pathways, EMT activation, poor prognosis, and drug resistance. Immunohistochemistry (IHC) results demonstrated that high CD8+ T cell infiltration was associated with low-CR-protein expression. Importantly, patients with low CRS were more likely to benefit from immune checkpoint blockade (ICB) treatment, possibly due to their higher tumor mutation burden (TMB), increased immune checkpoint expression, and higher proportion of “hot” immunophenotype.
Conclusion
In a nutshell, the cross talk in CR could reflect the TME immunoreactivity in breast cancer. Besides providing the first comprehensive pathway-level analysis of CR in breast cancer, this work highlights the potential clinical utility of CR for immunotherapy.







Similar content being viewed by others
Availability of data and materials
All online data and tools described in this article are available from their web servers and are free for any scientist to use for non-commercial purposes. Further information and source code are available from the corresponding author upon reasonable request.
Abbreviations
- BC:
-
Breast cancer
- CR:
-
Circadian rhythm
- CRS:
-
Circadian rhythm signature
- ER:
-
Estrogen receptor
- ICB:
-
Immune checkpoint blockade
- IPS:
-
Immunophenoscore
- TIDE:
-
Tumor Immune Dysfunction and Evolution
- TME:
-
Tumor microenvironment
- TMB:
-
Tumor mutation burden
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Shao D, Cheng S, Guo F, Zhu C, Yuan Y, Hu K et al (2020) Prevalence of hereditary breast and ovarian cancer (HBOC) predisposition gene mutations among 882 HBOC high-risk Chinese individuals. Cancer Sci 111:647–657. https://doi.org/10.1111/cas.14242
Hansen J (2017) Night Shift Work and Risk of Breast Cancer. Curr Environ Health Rep 4:325–339. https://doi.org/10.1007/s40572-017-0155-y
Srour B, Plancoulaine S, Andreeva VA, Fassier P, Julia C, Galan P et al (2018) Circadian nutritional behaviours and cancer risk: New insights from the NutriNet-santé prospective cohort study: Disclaimers. Int J Cancer 143:2369–2379. https://doi.org/10.1002/ijc.31584
Xiao Q, James P, Breheny P, Jia P, Park Y, Zhang D et al (2020) Outdoor light at night and postmenopausal breast cancer risk in the NIH-AARP diet and health study. Int J Cancer 147:2363–2372. https://doi.org/10.1002/ijc.33016
Scheiermann C, Kunisaki Y, Frenette PS (2013) Circadian control of the immune system. Nat Rev Immunol 13:190–198. https://doi.org/10.1038/nri3386
Collins EJ, Cervantes-Silva MP, Timmons GA, O’Siorain JR, Curtis AM, Hurley JM (2021) Post-transcriptional circadian regulation in macrophages organizes temporally distinct immunometabolic states. Genome Res. https://doi.org/10.1101/gr.263814.120
Hadadi E, Taylor W, Li X-M, Aslan Y, Villote M, Rivière J et al (2020) Chronic circadian disruption modulates breast cancer stemness and immune microenvironment to drive metastasis in mice. Nat Commun 11:3193. https://doi.org/10.1038/s41467-020-16890-6
Wu Y, Tao B, Zhang T, Fan Y, Mao R (2019) Pan-cancer analysis reveals disrupted circadian clock associates with T cell exhaustion. Front Immunol 10:2451. https://doi.org/10.3389/fimmu.2019.02451
Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T et al (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27:1160–1167. https://doi.org/10.1200/JCO.2008.18.1370
Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7. https://doi.org/10.1186/1471-2105-14-7
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420. https://doi.org/10.1038/nbt.4096
Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H et al (2021) Inference and analysis of cell-cell communication using Cell Chat. Nat Commun 12:1088. https://doi.org/10.1038/s41467-021-21246-9
Wang J, Vasaikar S, Shi Z, Greer M, Zhang B, WebGestalt, (2017) a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 45(2017):W130–W137. https://doi.org/10.1093/nar/gkx356
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W et al (2013) Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4:2612. https://doi.org/10.1038/ncomms3612
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018) Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 1711:243–259. https://doi.org/10.1007/978-1-4939-7493-1_12
Xu L, Deng C, Pang B, Zhang X, Liu W, Liao G et al (2018) TIP: a web server for resolving tumor immunophenotype profiling. Cancer Res 78:6575–6580. https://doi.org/10.1158/0008-5472.CAN-18-0689
Zeng D, Ye Z, Shen R, Yu G, Wu J, Xiong Y et al (2021) IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front Immunol 12:687975. https://doi.org/10.3389/fimmu.2021.687975
Ringel AE, Drijvers JM, Baker GJ, Catozzi A, García-Cañaveras JC, Gassaway BM et al (2020) Obesity shapes metabolism in the tumor microenvironment to suppress anti-tumor immunity. Cell 183:1848. https://doi.org/10.1016/j.cell.2020.11.009
Wilkerson MD, Hayes DN (2010) ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26:1572–1573. https://doi.org/10.1093/bioinformatics/btq170
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D et al (2017) Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 18:248–262. https://doi.org/10.1016/j.celrep.2016.12.019
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X et al (2018) Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 24:1550–1558. https://doi.org/10.1038/s41591-018-0136-1
Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y et al (2018) TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554:544–548. https://doi.org/10.1038/nature25501
Maeser D, Gruener RF, Huang RS (2021) oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform 22:6. https://doi.org/10.1093/bib/bbab260
Kareva I (2019) Metabolism and gut microbiota in cancer immunoediting, CD8/treg ratios, immune cell homeostasis, and cancer (immuno)therapy: concise review. Stem Cells 37:1273–1280. https://doi.org/10.1002/stem.3051
Wang C, Cao M, Jiang X, Yao Y, Liu Z, Luo D (2021) Macrophage balance fraction determines the degree of immunosuppression and metastatic ability of breast cancer. Int Immunopharmacol 97:107682. https://doi.org/10.1016/j.intimp.2021.107682
Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A et al (2021) A single-cell and spatially resolved atlas of human breast cancers. Nat Genet 53:1334–1347. https://doi.org/10.1038/s41588-021-00911-1
Lesicka M, Jabłońska E, Wieczorek E, Seroczyńska B, Siekierzycka A, Skokowski J et al (2018) Altered circadian genes expression in breast cancer tissue according to the clinical characteristics. PLoS ONE 13:e0199622
Davis K, Roden LC, Leaner VD, van der Watt PJ (2019) The tumour suppressing role of the circadian clock. IUBMB Life 71:771–780. https://doi.org/10.1002/iub.2005
Zeng Q, Michael IP, Zhang P, Saghafinia S, Knott G, Jiao W et al (2019) Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 573:526–531. https://doi.org/10.1038/s41586-019-1576-6
Tian R, Li Y, Shu M (2021) Circadian regulation patterns with distinct immune landscapes in gliomas aid in the development of a risk model to predict prognosis and therapeutic response. Front Immunol 12:797450. https://doi.org/10.3389/fimmu.2021.797450
Iurisci I, Filipski E, Reinhardt J, Bach S, Gianella-Borradori A, Iacobelli S et al (2006) Improved tumor control through circadian clock induction by Seliciclib, a cyclin-dependent kinase inhibitor. Cancer Res 66:10720–10728
Kiessling S, Beaulieu-Laroche L, Blum ID, Landgraf D, Welsh DK, Storch K-F et al (2017) Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol 15:13. https://doi.org/10.1186/s12915-017-0349-7
Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP (2007) The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene 26:7916–7920
Hill SM, Belancio VP, Dauchy RT, Xiang S, Brimer S, Mao L et al (2015) Melatonin: an inhibitor of breast cancer. Endocr Relat Cancer 22:R183–R204. https://doi.org/10.1530/ERC-15-0030
Martínez-Campa C, Alonso-González C, Mediavilla MD, Cos S, González A, Ramos S et al (2006) Melatonin inhibits both ER alpha activation and breast cancer cell proliferation induced by a metalloestrogen, cadmium. J Pineal Res 40:291–296
Semiglazova T, Tsyrlina Y, Poltoratskiy A, Busko Y, Apollonova V (2018) Melatonin and metformin in neoadjuvant hormonotherapy in locally advanced breast cancer. Vopr Onkol 64:612–619. https://doi.org/10.37469/0507-3758-2018-64-5-612-619
Yu C-W, Cheng K-C, Chen L-C, Lin M-X, Chang Y-C, Hwang-Verslues WW (1861) Pro-inflammatory cytokines IL-6 and CCL2 suppress expression of circadian gene Period2 in mammary epithelial cells. Biochim Biophys Acta Gene Regul Mech 2018:1007–1017. https://doi.org/10.1016/j.bbagrm.2018.09.003
Reeves E, James E (2017) Antigen processing and immune regulation in the response to tumours. Immunology 150:16–24. https://doi.org/10.1111/imm.12675
Herbelet S, De Bleecker JL (2018) Immune checkpoint failures in inflammatory myopathies: an overview. Autoimmun Rev 17:746–754. https://doi.org/10.1016/j.autrev.2018.01.026
Chabanon RM, Pedrero M, Lefebvre C, Marabelle A, Soria J-C, Postel-Vinay S (2016) Mutational Landscape and Sensitivity to Immune Checkpoint Blockers. Clin Cancer Res 22:4309–4321. https://doi.org/10.1158/1078-0432.CCR-16-0903
Maleki Vareki S (2018) High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer 6:157. https://doi.org/10.1186/s40425-018-0479-7
Paulson KG, Voillet V, McAfee MS, Hunter DS, Wagener FD, Perdicchio M et al (2018) Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun 9:3868. https://doi.org/10.1038/s41467-018-06300-3
Bevinakoppamath S, Ramachandra SC, Yadav AK, Basavaraj V, Vishwanath P, Prashant A (2021) Understanding the emerging link between circadian rhythm, Nrf2 pathway, and breast cancer to overcome drug resistance. Front Pharmacol 12:719631. https://doi.org/10.3389/fphar.2021.719631
Acknowledgements
The authors highly appreciate Gene Expression Omnibus (GEO) and the Cancer Genome Atlas (TCGA) database for providing the transcriptome and clinical information.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 81560464 and 31960152 (D.L.).
Author information
Authors and Affiliations
Contributions
SX performed the bioinformatic analysis, statistical analysis, and drafting of the manuscript. WZ performed the experiment. LW, TZ, WW, OZ, and XX contributed to the investigation, methodology, and data curation. ZL and DL conceived of the study, participated in its design, and revised it substantially. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethics approval and consent to participate
This study was approved by the institutional review board of the First Affiliated Hospital of Nanchang University, and written informed consent was obtained from all patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiong, S., Zhu, W., Wu, L. et al. Circadian pattern subtyping unveiling distinct immune landscapes in breast cancer patients for better immunotherapy. Cancer Immunol Immunother 72, 3293–3307 (2023). https://doi.org/10.1007/s00262-023-03495-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-023-03495-3