Abstract
Background
Chimeric antigen receptor-modified T cells (CAR T-cells) have shown exhilarative clinical efficacy for hematological malignancies. However, a shared antigen pool between healthy and malignant T-cells remains a concept to be technically and clinically explored for CAR T-cell therapy in T-cell cancers. No guidelines for engineering CAR T-cells targeting self-expressed antigens are currently available.
Method
Based on anti-CD70 CAR (CAR-70) T-cells, we constructed CD70 knock-out and wild-type CAR (CAR-70KO and CAR-70WT) T-cells and evaluated their manufacturing and anti-tumor capability. Single-cell RNA sequencing and TCR sequencing were performed to further reveal the underlying differences between the two groups of CAR T-cells.
Results
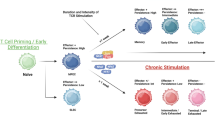
Our data showed that the disruption of target genes in T-cells before CAR transduction advantaged the expansion and cell viability of CAR T-cells during manufacturing periods, as well as the degranulation, anti-tumor efficacy, and proliferation potency in response to tumor cells. Meanwhile, more naïve and central memory phenotype CAR+ T-cells, with higher TCR clonal diversity, remained in the final products in KO samples. Gene expression profiles revealed a higher activation and exhaustion level of CAR-70WT T-cells, while signaling transduction pathway analysis identified a higher level of the phosphorylation-related pathway in CAR-70KO T-cells.
Conclusion
This study evidenced that CD70 stimulation during manufacturing process induced early exhaustion of CAR-70 T-cells. Knocking-out CD70 in T-cells prevented the exhaustion and led to a better-quality CAR-70 T-cell product. Our research will contribute to good engineering CAR T-cells targeting self-expressed antigens.





Similar content being viewed by others
Availability of data and materials
Data were contained in the manuscript, and raw data for the scRNA-seq and scTCR-seq are available via HRA002418 at the National Genome Data Center (NGDC) (https://ngdc.cncb.ac.cn/).
References
Singh AK, McGuirk JP (2020) CAR T cells: continuation in a revolution of immunotherapy. Lancet Oncol 21(3):e168–e178
Lu J, Jiang G (2022) The journey of CAR-T therapy in hematological malignancies. Mol Cancer 21(1):194
Wang N, Hu X, Cao W, Li C, Xiao Y, Cao Y et al (2020) Efficacy and safety of CAR19/22 T-cell cocktail therapy in patients with refractory/relapsed B-cell malignancies. Blood 135(1):17–27
Wang D, Wang J, Hu G, Wang W, Xiao Y, Cai H et al (2021) A phase 1 study of a novel fully human BCMA-targeting CAR (CT103A) in patients with relapsed/refractory multiple myeloma. Blood 137(21):2890–2901
Safarzadeh Kozani P, Safarzadeh Kozani P, Rahbarizadeh F (2021) CAR-T cell therapy in T-cell malignancies: Is success a low-hanging fruit? Stem Cell Res Ther 12(1):527
Hamieh M, Dobrin A, Cabriolu A, van der Stegen SJC, Giavridis T, Mansilla-Soto J et al (2019) CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature 568(7750):112–116
Zhou S, Zhu X, Shen N, Li Q, Wang N, You Y et al (2019) T cells expressing CD26-specific chimeric antigen receptors exhibit extensive self-antigen-driven fratricide. Immunopharmacol Immunotoxicol 41(4):490–496
Jacobs J, Deschoolmeester V, Zwaenepoel K, Rolfo C, Silence K, Rottey S et al (2015) CD70: an emerging target in cancer immunotherapy. Pharmacol Ther 155:1–10
Flieswasser T, Camara-Clayette V, Danu A, Bosq J, Ribrag V, Zabrocki P et al (2019) Screening a broad range of solid and haematological tumour types for CD70 expression using a uniform IHC methodology as potential patient stratification method. Cancers 11(10):1611
Park YP, Jin L, Bennett KB, Wang D, Fredenburg KM, Tseng JE et al (2018) CD70 as a target for chimeric antigen receptor T cells in head and neck squamous cell carcinoma. Oral Oncol 78:145–150
Panowski SH, Srinivasan S, Tan N, Tacheva-Grigorova SK, Smith B, Mak YSL et al (2022) Preclinical development and evaluation of allogeneic CAR T cells targeting CD70 for the treatment of renal cell carcinoma. Can Res 82(14):2610–2624
Leick MB, Silva H, Scarfò I, Larson R, Choi BD, Bouffard AA et al (2022) Non-cleavable hinge enhances avidity and expansion of CAR-T cells for acute myeloid leukemia. Cancer Cell 40(5):494-508.e495
Sauer T, Parikh K, Sharma S, Omer B, Sedloev D, Chen Q et al (2021) CD70-specific CAR T cells have potent activity against acute myeloid leukemia without HSC toxicity. Blood 138(4):318–330
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36(5):411–420
McGinnis CS, Murrow LM, Gartner ZJ (2019) DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 8(4):329-337.e324
Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ et al (2016) Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352(6282):189–196
Borcherding N, Bormann NL, Kraus G (2020) scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Research 9:47
Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q et al (2018) Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564(7735):268–272
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S et al (2021) The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49(D1):D605-d612
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA et al (2017) Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med 377(26):2531–2544
Alcantara M, Tesio M, June CH, Houot R (2018) CAR T-cells for T-cell malignancies: challenges in distinguishing between therapeutic, normal, and neoplastic T-cells. Leukemia 32(11):2307–2315
Went P, Agostinelli C, Gallamini A, Piccaluga PP, Ascani S, Sabattini E et al (2006) Marker expression in peripheral T-cell lymphoma: a proposed clinical-pathologic prognostic score. J Clin Oncol 24(16):2472–2479
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157(6):1262–1278
Pal SK, Forero-Torres A, Thompson JA, Morris JC, Chhabra S, Hoimes CJ et al (2019) A phase 1 trial of SGN-CD70A in patients with CD70-positive, metastatic renal cell carcinoma. Cancer 125(7):1124–1132
Riether C, Pabst T, Höpner S, Bacher U, Hinterbrandner M, Banz Y et al (2020) Targeting CD70 with cusatuzumab eliminates acute myeloid leukemia stem cells in patients treated with hypomethylating agents. Nat Med 26(9):1459–1467
Deng W, Chen P, Lei W, Xu Y, Xu N, Pu JJ et al (2021) CD70-targeting CAR-T cells have potential activity against CD19-negative B-cell lymphoma. Cancer Commun (London, England) 41(9):925–929
Yang M, Tang X, Zhang Z, Gu L, Wei H, Zhao S et al (2020) Tandem CAR-T cells targeting CD70 and B7–H3 exhibit potent preclinical activity against multiple solid tumors. Theranostics 10(17):7622–7634
Gomes-Silva D, Srinivasan M, Sharma S, Lee CM, Wagner DL, Davis TH et al (2017) CD7-edited T cells expressing a CD7-specific CAR for the therapy of T-cell malignancies. Blood 130(3):285–296
Freiwan A, Zoine J, Crawford JC, Vaidya A, Schattgen SA, Myers J et al (2022) Engineering naturally occurring CD7 negative T cells for the Immunotherapy of hematological malignancies. Blood 25:2684–2696
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH (2015) Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 4(11):e264
Borst J, Hendriks J, Xiao Y (2005) CD27 and CD70 in T cell and B cell activation. Curr Opin Immunol 17(3):275–281
McLellan AD, Ali Hosseini Rad SM (2019) Chimeric antigen receptor T cell persistence and memory cell formation. Immunol Cell Biol 97(7):664–674
Xu Y, Zhang M, Ramos CA, Durett A, Liu E, Dakhova O et al (2014) Closely related T-memory stem cells correlate with in vivo expansion of CAR.CD19-T cells and are preserved by IL-7 and IL-15. Blood 123(24):3750–3759
Gaud G, Lesourne R, Love PE (2018) Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol 18(8):485–497
Salter AI, Ivey RG, Kennedy JJ, Voillet V, Rajan A, Alderman EJ et al (2018) Phosphoproteomic analysis of chimeric antigen receptor signaling reveals kinetic and quantitative differences that affect cell function. Sci Signal. https://doi.org/10.1126/scisignal.aat6753
Acknowledgements
We gratefully thank Wei Mu for the instructions for manufacturing the CAR T-cells.
Funding
This work was supported by the National Key R&D Program of China (2021YFF0703704), National Natural Science Foundation of China (82270183, 32100527, and 82100241), Natural Sciences Foundation of Hubei Province of China (2020CFB790), and the Excellent Young Science Foundation Project of Tongji Hospital (No.2020YQ0012).
Author information
Authors and Affiliations
Contributions
A-YG, QL and XZ proposed the ideas, oversaw the whole project, and revised the manuscript. JC conducted the functional experiments and analyzed the related data. Y. Zhao and HH analyzed the data of scRNA and scTCR sequencing. LT and SD helped with the experiments and participated in the discussion. JC, Y. Zhao and Y. Zeng wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
There is no competing interest for all the authors.
Ethics approval
Not applicable.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, J., Zhao, Y., Hu, H. et al. Revealing the impact of CD70 expression on the manufacture and functions of CAR-70 T-cells based on single-cell transcriptomics. Cancer Immunol Immunother 72, 3163–3174 (2023). https://doi.org/10.1007/s00262-023-03475-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-023-03475-7