Abstract
Background
Immune checkpoint inhibitors (ICIs) have dramatically changed the landscape of cancer treatment. However, only a few patients respond to ICI treatment. Thus, uncovering clinically accessible ICI biomarkers would help identify which patients will respond well to ICI treatment. A comprehensive objective response rate (ORR) data of anti-PD-1/PD-L1 monotherapy in pan-cancer would offer the original data to explore the new biomarkers for ICIs.
Methods
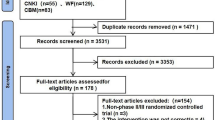
We systematically searched PubMed, Cochrane, and Embase for clinical trials on July 1, 2021, limited to the years 2017–2021, from which we obtained studies centering around anti–PD-1/PD-L1 monotherapy. Finally, 121 out of 3099 publications and 143 ORR data were included. All of the 31 tumor types/subtypes can be found in the TCGA database. The gene expression profiles and mutation data were downloaded from TCGA. A comprehensive genome-wide screening of ORR highly correlated mutations among 31 cancers was conducted by Pearson correlation analysis based on the TCGA database.
Results
According to the ORR, we classified 31 types of cancer into high, medium, and low response types. Further analysis uncovered that “high response” cancers had more T cell infiltration, more neoantigens, and less M2 macrophage infiltration. A panel of 28 biomarkers reviewed from recent articles were investigated with ORR. We also found the TMB as a traditional biomarker had a high correlation coefficient with ORR in pan-cancer, however, the correlation between ITH and ORR was low across pan-cancer. Moreover, we primarily identified 1044 ORR highly correlated mutations through a comprehensive screening of TCGA data, among which USH2A, ZFHX4 and PLCO mutations were found to be highly correlated to strengthened tumor immunogenicity and inflamed antitumor immunity, as well as improved outcomes for ICIs treatment among multiple immunotherapy cohorts.
Conclusion
Our study provides comprehensive data on ORR of anti-PD-1/PD-L1 monotherapy across 31 tumor types/subtypes and an essential reference of ORR to explore new biomarkers. We also screened out a list of 1044 immune response related genes and we showed that USH2A, ZFHX4 and PLCO mutations may act as good biomarkers for predicting patient response to anti-PD-1/PD-L1 ICIs.






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CYT score:
-
Cytolytic activity score
- ICIs:
-
Immune checkpoint inhibitors
- IRRG:
-
Immune response related genes
- ITH:
-
Intratumoral heterogeneity
- NSCLC:
-
Non-small cell lung cancer
- ORR:
-
Objective response rate
- OS:
-
Overall survival
- PD-1/PD-L1:
-
Protein programmed cell death protein 1/protein programmed death receptor ligand-1
- PFS:
-
Progression-free survival
- TIL:
-
Tumor-infiltrating lymphocyte
- TMB:
-
Tumor mutational burden
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics. CA a Cancer J Clin 71:7–33. https://doi.org/10.3322/caac.21654
Ribas A, Wolchok JD (2018) Cancer immunotherapy using checkpoint blockade. Science 359:1350–1355. https://doi.org/10.1126/science.aar4060
Zhou X, Yao Z, Bai H et al (2021) Treatment-related adverse events of PD-1 and PD-L1 inhibitor-based combination therapies in clinical trials: a systematic review and meta-analysis. Lancet Oncol 22:1265–1274. https://doi.org/10.1016/S1470-2045(21)00333-8
Samstein RM, Lee CH, Shoushtari AN et al (2019) Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 51:202–206. https://doi.org/10.1038/s41588-018-0312-8
Litchfield K, Reading JL, Puttick C et al (2021) Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell. 184:596–614. https://doi.org/10.1016/j.cell.2021.01.002
Miao D, Margolis CA, Vokes NI et al (2018) Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet 50:1271–1281. https://doi.org/10.1038/s41588-018-0200-2
Morad G, Helmink BA, Sharma P, Wargo JA (2021) Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 184:5309–5337. https://doi.org/10.1016/j.cell.2021.09.020
Liu D, Schilling B, Liu D et al (2019) Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 25:1916–1927. https://doi.org/10.1038/s41591-019-0654-5
Riaz N, Havel JJ, Makarov V et al (2017) Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 171:934–49. https://doi.org/10.1016/j.cell.2017.09.028
Snyder A, Makarov V, Merghoub T et al (2014) Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371:2189–2199. https://doi.org/10.1056/NEJMoa1406498
Van Allen EM, Miao D, Schilling B et al (2015) Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350:207–211. https://doi.org/10.1126/science.aad0095
Hellmann MD, Nathanson T, Rizvi H et al (2018) Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 33:843–52. https://doi.org/10.1016/j.ccell.2018.03.018
Rizvi NA, Hellmann MD, Snyder A et al (2015) Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348:124–128. https://doi.org/10.1126/science.aaa1348
Yarchoan M, Hopkins A, Jaffee EM (2017) Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 377:2500–2501. https://doi.org/10.1056/NEJMc1713444
Hoadley KA, Yau C, Hinoue T et al (2018) Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173:291–304. https://doi.org/10.1016/j.cell.2018.03.022
Liu Y, Sethi NS, Hinoue T et al (2018) Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 33:721–35. https://doi.org/10.1016/j.ccell.2018.03.010
Tumeh PC, Harview CL, Yearley JH et al (2014) PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515:568–571. https://doi.org/10.1038/nature13954
Chow MT, Ozga AJ, Servis RL, Frederick DT, Lo JA, Fisher DE, Freeman GJ, Boland GM, Luster AD (2019) Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity. 50:1498–512. https://doi.org/10.1016/j.immuni.2019.04.010
Gibney GT, Weiner LM, Atkins MB (2016) Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol 17:e542–e551. https://doi.org/10.1016/S1470-2045(16)30406-5
Chen L, Diao L, Yang Y et al (2018) CD38-mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 blockade. Cancer Discov 8:1156–1175. https://doi.org/10.1158/2159-8290.CD-17-1033
Koyama S, Akbay EA, Li YY et al (2016) Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun 7:10501. https://doi.org/10.1038/ncomms10501
Huang L, Malu S, McKenzie JA et al (2018) The RNA-binding Protein MEX3B mediates resistance to cancer immunotherapy by downregulating HLA-A expression. Clin Cancer Res 24:3366–3376. https://doi.org/10.1158/1078-0432.CCR-17-2483
Wolf Y, Bartok O, Patkar S et al (2019) UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell 179:219–35. https://doi.org/10.1016/j.cell.2019.08.032
Wang L, Saci A, Szabo PM et al (2018) EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun 9:3503. https://doi.org/10.1038/s41467-018-05992-x
Ayers M, Lunceford J, Nebozhyn M et al (2017) IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 127:2930–2940. https://doi.org/10.1172/JCI91190
Mariathasan S, Turley SJ, Nickles D et al (2018) TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554:544–548. https://doi.org/10.1038/nature25501
Pabla S, Conroy JM, Nesline MK et al (2019) Proliferative potential and resistance to immune checkpoint blockade in lung cancer patients. J Immunother Cancer 7:27. https://doi.org/10.1186/s40425-019-0506-3
Messina JL, Fenstermacher DA, Eschrich S et al (2012) 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci Rep 2:765. https://doi.org/10.1038/srep00765
Tokunaga R, Nakagawa S, Sakamoto Y et al (2020) 12-Chemokine signature, a predictor of tumor recurrence in colorectal cancer. Int J Cancer 147:532–541. https://doi.org/10.1002/ijc.32982
McDermott DF, Huseni MA, Atkins MB et al (2018) Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 24:749–757. https://doi.org/10.1038/s41591-018-0053-3
Fehrenbacher L, Spira A, Ballinger M et al (2016) Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387:1837–1846. https://doi.org/10.1016/S0140-6736(16)00587-0
Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N (2015) Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160:48–61. https://doi.org/10.1016/j.cell.2014.12.033
Shin DS, Zaretsky JM, Escuin-Ordinas H et al (2017) Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov 7:188–201. https://doi.org/10.1158/2159-8290.CD-16-1223
Riaz N, Havel JJ, Kendall SM, Makarov V, Walsh LA, Desrichard A, Weinhold N, Chan TA (2016) Recurrent SERPINB3 and SERPINB4 mutations in patients who respond to anti-CTLA4 immunotherapy. Nat Genet 48:1327–1329. https://doi.org/10.1038/ng.3677
Conway JR, Kofman E, Mo SS, Elmarakeby H, Van Allen E (2018) Genomics of response to immune checkpoint therapies for cancer: implications for precision medicine. Genome Med 10:93. https://doi.org/10.1186/s13073-018-0605-7
Shrestha R, Nabavi N, Lin YY et al (2019) BAP1 haploinsufficiency predicts a distinct immunogenic class of malignant peritoneal mesothelioma. Genome Med 11:8. https://doi.org/10.1186/s13073-019-0620-3
Peng W, Chen JQ, Liu C et al (2016) Loss of PTEN promotes resistance to T Cell-mediated immunotherapy. Cancer Discov 6:202–216. https://doi.org/10.1158/2159-8290.CD-15-0283
Aredo JV, Padda SK, Kunder CA, Han SS, Neal JW, Shrager JB, Wakelee HA (2019) Impact of KRAS mutation subtype and concurrent pathogenic mutations on non-small cell lung cancer outcomes. Lung Cancer 133:144–150. https://doi.org/10.1016/j.lungcan.2019.05.015
Martin TD, Patel RS, Cook DR, Choi MY, Patil A, Liang AC, Li MZ, Haigis KM, Elledge SJ (2021) The adaptive immune system is a major driver of selection for tumor suppressor gene inactivation. Science 373:1327–1335. https://doi.org/10.1126/science.abg5784
Gettinger S, Choi J, Hastings K et al (2017) Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov 7:1420–1435. https://doi.org/10.1158/2159-8290.CD-17-0593
Castro A, Ozturk K, Pyke RM, Xian S, Zanetti M, Carter H (2019) Elevated neoantigen levels in tumors with somatic mutations in the HLA-A, HLA-B, HLA-C and B2M genes. BMC Med Genom 12:107. https://doi.org/10.1186/s12920-019-0544-1
Colaprico A, Silva TC, Olsen C et al (2016) TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 44:e71. https://doi.org/10.1093/nar/gkv1507
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Sen T, Rodriguez BL, Chen L et al (2019) Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov 9:646–661. https://doi.org/10.1158/2159-8290.CD-18-1020
Eroglu Z, Zaretsky JM, Hu-Lieskovan S et al (2018) High response rate to PD-1 blockade in desmoplastic melanomas. Nature 553:347–350. https://doi.org/10.1038/nature25187
Wang G, Chow RD, Zhu L et al (2020) CRISPR-GEMM pooled mutagenic screening identifies KMT2D as a major modulator of immune checkpoint blockade. Cancer Discov 10:1912–1933. https://doi.org/10.1158/2159-8290.CD-19-1448
Xie X, Tang Y, Sheng J, Shu P, Zhu X, Cai X, Zhao C, Wang L, Huang X (2021) Titin mutation is associated with tumor mutation burden and promotes antitumor immunity in lung squamous cell carcinoma. Front Cell Dev Biol. 9:761758. https://doi.org/10.3389/fcell.2021.761758
Li X, Pasche B, Zhang W, Chen K (2018) Association of MUC16 mutation with tumor mutation load and outcomes in patients with gastric cancer. JAMA Oncol 4:1691–1698. https://doi.org/10.1001/jamaoncol.2018.2805
Brown LC, Tucker MD, Sedhom R et al (2021) LRP1B mutations are associated with favorable outcomes to immune checkpoint inhibitors across multiple cancer types. J Immunother Cancer. https://doi.org/10.1136/jitc-2020-001792
Li P, Xiao J, Zhou B, Wei J, Luo J, Chen W (2020) SYNE1 mutation may enhance the response to immune checkpoint blockade therapy in clear cell renal cell carcinoma patients. Aging (Albany NY) 12:19316–24. https://doi.org/10.18632/aging.103781
Jhunjhunwala S, Hammer C, Delamarre L (2021) Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer 21:298–312. https://doi.org/10.1038/s41568-021-00339-z
Bockorny B, Semenisty V, Macarulla T et al (2020) BL-8040, a CXCR4 antagonist, in combination with pembrolizumab and chemotherapy for pancreatic cancer: the COMBAT trial. Nat Med 26:878–885. https://doi.org/10.1038/s41591-020-0880-x
Lussier DM, Alspach E, Ward JP et al (2021) Radiation-induced neoantigens broaden the immunotherapeutic window of cancers with low mutational loads. Proc Natl Acad Sci U S A 118:e2102611118. https://doi.org/10.1073/pnas.2102611118
Zhu X, Cao Y, Liu W, Ju X, Zhao X, Jiang L, Ye Y, Jin G, Zhang H (2021) Stereotactic body radiotherapy plus pembrolizumab and trametinib versus stereotactic body radiotherapy plus gemcitabine for locally recurrent pancreatic cancer after surgical resection: an open-label, randomised, controlled, phase 2 trial. Lancet Oncol 22:1093–1102. https://doi.org/10.1016/S1470-2045(21)00286-2
Zhang Y, Chen H, Mo H et al (2021) Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell. https://doi.org/10.1016/j.ccell.2021.09.010
Anderson NR, Minutolo NG, Gill S, Klichinsky M (2021) Macrophage-based approaches for cancer immunotherapy. Cancer Res 81:1201–1208. https://doi.org/10.1158/0008-5472.CAN-20-2990
Li J, Wang W, Zhang Y et al (2020) Epigenetic driver mutations in ARID1A shape cancer immune phenotype and immunotherapy. J Clin Invest 130:2712–2726. https://doi.org/10.1172/JCI134402
Jia Q, Wang J, He N, He J, Zhu B (2019) Titin mutation associated with responsiveness to checkpoint blockades in solid tumors. JCI Insight. https://doi.org/10.1172/jci.insight.127901
Acknowledgements
None.
Funding
This work was funded by the Public Welfare Technology Application Research Project of Zhejiang Province (No. LGF22H200013) and the Zhejiang Provincial Medical and Health Science and Technology Project (No. 2022KY1228).
Author information
Authors and Affiliations
Contributions
Conceptualization, Methodology, Formal analysis: YM, WW, HX, ML, SL, LZ, FG. Software, Data Curation: YM, WW, HX, ML. Writing- Original draft preparation: YM, HX, ML, QY, ZS. Writing–Reviewing and Editing: YM, HX, ML, QY, ZS, LZ, FG, SL, WW. Visualization, Supervision: YM, WW, SL, LZ and FG. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflict of interests to declare.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All the genetic data and patient cohort information we used were publicly available datasets.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mao, Y., Xie, H., Lv, M. et al. The landscape of objective response rate of anti-PD-1/L1 monotherapy across 31 types of cancer: a system review and novel biomarker investigating. Cancer Immunol Immunother 72, 2483–2498 (2023). https://doi.org/10.1007/s00262-023-03441-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-023-03441-3