Abstract
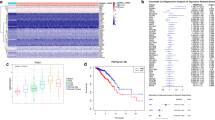
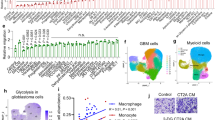
Globally, breast cancer is one of the leading causes of cancer death in women. Metabolic reprogramming and immune escape are two important mechanisms supporting the progression of breast cancer. Lactate in tumors mainly comes from glycolysis and glutaminolysis. Using multiomics data analysis, we found that lactate is mainly derived from glycolysis in breast cancer. Single-cell transcriptome analysis found that breast cancer cells with higher malignancy, especially those in the cell cycle, have higher expression levels of glycolytic metabolic enzymes. Combined with clinical data analysis, it was found that the expression of the lactate transporter SLC16A3 is correlated with breast cancer molecular subtypes and immune infiltration. Among 22 immune cells, macrophages are the most abundant immune cells in breast cancer tissues, and the proportion of M1 macrophages is lower in the high SLC16A3 expression group. Finally, in vitro experiments confirmed that lactate could inhibit the expression of M1 macrophage markers at both RNA and protein levels. In conclusion, we found that lactate produced by glycolysis regulates the polarization of inflammatory macrophages in breast cancer.








Similar content being viewed by others
Data availability
All the data in our study are available upon request.
References
Britt KL, Cuzick J, Phillips KA (2020) Key steps for effective breast cancer prevention. Nat Rev Cancer 20(8):417–436
Waks AG, Winer EP (2019) Breast cancer treatment: a review. JAMA 321(3):288–300
Dias AS, Almeida CR, Helguero LA, Duarte IF (2019) Metabolic crosstalk in the breast cancer microenvironment. Eur J Cancer 121:154–171
Zaslona Z, O’Neill LAJ (2020) Cytokine-like roles for metabolites in immunity. Mol Cell 78(5):814–823
Koppenol WH, Bounds PL, Dang CV (2011) Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer 11(5):325–337
Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB (2008) Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev 18(1):54–61
de la Cruz-Lopez KG, Castro-Munoz LJ, Reyes-Hernandez DO, Garcia-Carranca A, Manzo-Merino J (2019) Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front Oncol 9:1143
Certo M, Tsai CH, Pucino V, Ho PC, Mauro C (2021) Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat Rev Immunol 21(3):151–161
Gottfried E, Kunz-Schughart LA, Ebner S, Mueller-Klieser W, Hoves S, Andreesen R, Mackensen A, Kreutz M (2006) Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood 107(5):2013–2021
Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves S et al (2007) Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 109(9):3812–3819
Goetze K, Walenta S, Ksiazkiewicz M, Kunz-Schughart LA, Mueller-Klieser W (2011) Lactate enhances motility of tumor cells and inhibits monocyte migration and cytokine release. Int J Oncol 39(2):453–463
Cassetta L, Pollard JW (2018) Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov 17(12):887–904
Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T et al (2014) Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41(1):14–20
Wang C, Cao M, Jiang X, Yao Y, Liu Z, Luo D (2021) Macrophage balance fraction determines the degree of immunosuppression and metastatic ability of breast cancer. Int Immunopharmacol 97:107682
Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM et al (2014) Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513(7519):559–563
Chen P, Zuo H, Xiong H, Kolar MJ, Chu Q, Saghatelian A, Siegwart DJ, Wan Y (2017) Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis. Proc Natl Acad Sci U S A 114(3):580–585
Mu X, Shi W, Xu Y, Xu C, Zhao T, Geng B, Yang J, Pan J, Hu S, Zhang C et al (2018) Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle 17(4):428–438
Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M et al (2019) Metabolic regulation of gene expression by histone lactylation. Nature 574(7779):575–580
Vadevoo SMP, Gunassekaran GR, Lee C, Lee N, Lee J, Chae S, Park J-Y, Koo J, Lee B (2021) The macrophage odorant receptor Olfr78 mediates the lactate-induced M2 phenotype of tumor-associated macrophages. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.2102434118
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M (2021) KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 49(D1):D545–D551
Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, Griss J, Sevilla C, Matthews L, Gong C et al (2022) The reactome pathway knowledgebase 2022. Nucleic Acids Res 50(D1):D687–D692
Mueckler M, Thorens B (2013) The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med 34(2–3):121–138
Chen LQ, Cheung LS, Feng L, Tanner W, Frommer WB (2015) Transport of sugars. Annu Rev Biochem 84:865–894
Payen VL, Mina E, Van Hee VF, Porporato PE, Sonveaux P (2020) Monocarboxylate transporters in cancer. Mol Metab 33:48–66
Felmlee MA, Jones RS, Rodriguez-Cruz V, Follman KE, Morris ME (2020) Monocarboxylate transporters (SLC16): function, regulation, and role in health and disease. Pharmacol Rev 72(2):466–485
Najafi M, Hashemi Goradel N, Farhood B, Salehi E, Nashtaei MS, Khanlarkhani N, Khezri Z, Majidpoor J, Abouzaripour M, Habibi M et al (2019) Macrophage polarity in cancer: a review. J Cell Biochem 120(3):2756–2765
Terunuma A, Putluri N, Mishra P, Mathe EA, Dorsey TH, Yi M, Wallace TA, Issaq HJ, Zhou M, Killian JK et al (2014) MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest 124(1):398–412
Weinstein JN, Collisson EA, Mills GB, Mills KR, Shaw BA, Ozenberger KE, Shmulevich I, Sander C, Stuart JM (2013) The cancer genome atlas pan-cancer analysis project. Nat Genet 45(10):1113–1120. https://doi.org/10.1038/ng.2764
Gao GF, Parker JS, Reynolds SM, Silva TC, Wang L-B, Zhou W, Akbani R, Bailey M, Balu S, Berman BP et al (2019) Before and after: comparison of legacy and harmonized TCGA genomic data commons’ data. Cell Syst 9(1):24-34.e10. https://doi.org/10.1016/j.cels.2019.06.006
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN et al (2020) Visualizing and interpreting cancer genomics data via the xena platform. Nat Biotechnol 38(6):675–678
Cancer Genome Atlas N (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70. https://doi.org/10.1038/nature11412
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Benjamin Gross S, Sumer O, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. https://doi.org/10.1126/scisignal.2004088
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y et al (2012) The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 486(7403):346–352
Gendoo DM, Ratanasirigulchai N, Schroder MS, Pare L, Parker JS, Prat A, Haibe-Kains B (2016) Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 32(7):1097–1099
Gong TQ, Jiang YZ, Shao C, Peng WT, Liu MW, Li DQ, Zhang BY, Du P, Huang Y, Li FF et al (2022) Proteome-centric cross-omics characterization and integrated network analyses of triple-negative breast cancer. Cell Rep 38(9):110460
Ghergurovich JM, Lang JD, Levin MK, Briones N, Facista SJ, Mueller C, Cowan AJ, McBride MJ, Rodriguez ESR, Killian A et al (2021) Local production of lactate, ribose phosphate, and amino acids within human triple-negative breast cancer. Med (N Y) 2(6):736–754
Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, Thennavan A, Wang C, Torpy JR, Bartonicek N et al (2021) A single-cell and spatially resolved atlas of human breast cancers. Nat Genet 53(9):1334–1347
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M et al (2012) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41(D1):D991–D995. https://doi.org/10.1093/nar/gks1193
Xu M, Wang X, Li Y, Geng X, Jia X, Zhang L, Yang H (2021) Arachidonic acid metabolism controls macrophage alternative activation through regulating oxidative phosphorylation in ppargamma dependent manner. Front Immunol 12:618501
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10(1):1523
Hanzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14:7
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P (2015) The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 1(6):417–425
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36(5):411–420
Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA et al (2013) Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4:2612
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018) Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 1711:243–259
Calvo MB, Figueroa A, Pulido EG, Campelo RG, Aparicio LA (2010) Potential role of sugar transporters in cancer and their relationship with anticancer therapy. Int J Endocrinol 2010:1–14. https://doi.org/10.1155/2010/205357
Ancey PB, Contat C, Meylan E (2018) Glucose transporters in cancer-from tumor cells to the tumor microenvironment. FEBS J 285(16):2926–2943
Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23(1):27–47
Pavlova NN, Zhu J, Thompson CB (2022) The hallmarks of cancer metabolism: still emerging. Cell Metab 34(3):355–377
Liu Y, Beyer A, Aebersold R (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165(3):535–550
Jang C, Chen L, Rabinowitz JD (2018) Metabolomics and isotope tracing. Cell 173(4):822–837
Chung W, Eum HH, Lee HO, Lee KM, Lee HB, Kim KT, Ryu HS, Kim S, Lee JE, Park YH et al (2017) Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun 8:15081
Lee JS, Adler L, Karathia H, Carmel N, Rabinovich S, Auslander N, Keshet R, Stettner N, Silberman A, Agemy L et al (2018) Urea cycle dysregulation generates clinically relevant genomic and biochemical signatures. Cell 174(6):1559-1570.e22. https://doi.org/10.1016/j.cell.2018.07.019
Aikun F, Yao B, Dong T, Chen Y, Jia Yao Y, Liu HL, Bai H, Liu X, Zhang Y et al (2022) Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 185(8):1356-1372.e26. https://doi.org/10.1016/j.cell.2022.02.027
Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF et al (2008) Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest 118(12):3930–3942
Ho J, de Moura MB, Lin Y, Vincent G, Thorne S, Duncan LM, Hui-Min L, Kirkwood JM, Becker D, Van Houten B et al (2012) Importance of glycolysis and oxidative phosphorylation in advanced melanoma. Mol Cancer 11:76
Pisarsky L, Bill R, Fagiani E, Dimeloe S, Goosen RW, Hagmann J, Hess C, Christofori G (2016) Targeting metabolic symbiosis to overcome resistance to anti-angiogenic therapy. Cell Rep 15(6):1161–1174
Allen E, Mieville P, Warren CM, Saghafinia S, Li L, Peng MW, Hanahan D (2016) Metabolic symbiosis enables adaptive resistance to anti-angiogenic therapy that is dependent on mTOR signaling. Cell Rep 15(6):1144–1160
Jimenez-Valerio G, Martinez-Lozano M, Bassani N, Vidal A, Ochoa-de-Olza M, Suarez C, Garcia-Del-Muro X, Carles J, Vinals F, Graupera M et al (2016) Resistance to antiangiogenic therapies by metabolic symbiosis in renal cell carcinoma PDX models and patients. Cell Rep 15(6):1134–1143
Lin S, Sun L, Lyu X, Ai X, Du D, Su N, Li H, Zhang L, Yu J, Yuan S (2017) Lactate-activated macrophages induced aerobic glycolysis and epithelial-mesenchymal transition in breast cancer by regulation of CCL5-CCR5 axis: a positive metabolic feedback loop. Oncotarget 8(66):110426–110443
Irizarry-Caro RA, McDaniel MM, Overcast GR, Jain VG, Troutman TD, Pasare C (2020) TLR signaling adapter BCAP regulates inflammatory to reparatory macrophage transition by promoting histone lactylation. Proc Natl Acad Sci U S A 117(48):30628–30638
Cui H, Xie N, Banerjee S, Ge J, Jiang D, Dey T, Matthews QL, Liu RM, Liu G (2021) Lung Myofibroblasts promote macrophage profibrotic activity through lactate-induced histone lactylation. Am J Respir Cell Mol Biol 64(1):115–125
Acknowledgements
The authors would like to thank Mrs. Zhuoqi Liu and Mrs. Xiaohong Yang for their insightful comments during their review of this manuscript.
Funding
This work was supported by National Natural Science Foundation of China (81560464 and 31960152 to D.L.), Applied Research and Cultivation Program of Jiangxi Provincial Department of Science and Technology (20212BAG70036 to S.Z.) and Jiangxi Provincial Education Department foundation Project (GJJ218911 to S.Z.).
Author information
Authors and Affiliations
Contributions
CW contributed to Conceptualization, Methodology, Software, Validation, Visualization, Formal analysis, Investigation, Data Curation, Writing-Original Draft; LX contributed to Methodology; Validation; Resources; Writing-Review and Editing; WZ contributed to Methodology; Validation; Resources; LL: Validation; Resources; SZ contributed to Conceptualization, Writing-Review and Editing; DL contributed to Conceptualization, Resources, Writing-Review and Editing, Supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, C., Xue, L., Zhu, W. et al. Lactate from glycolysis regulates inflammatory macrophage polarization in breast cancer. Cancer Immunol Immunother 72, 1917–1932 (2023). https://doi.org/10.1007/s00262-023-03382-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-023-03382-x