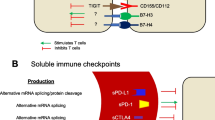
Abstract
Immune checkpoint blockade (ICB) of the programmed cell death 1/programmed cell death ligand 1 (PD-1/PD-L1) immune checkpoint pathway has led to unprecedented advances in cancer therapy. However, the overall response rate of anti-PD-1/PD-L1 monotherapy is still unpromising, underscoring the need for predictive biomarkers. In this retrospective study, we collected pretreatment plasma samples from two independent cohorts of patients receiving ICB. To determine whether a signature of plasma cytokines could be associated with therapeutic efficacy, we systemically profiled cytokine clusters and functional groups in the discovery and validation datasets by using 59 multiplexed bead immunoassays and bioinformatics analysis. We first attempted to functionally classify the 59 immunological factors according to their biological classification or functional roles in the cancer-immunity cycle. Surprisingly, we observed that two signatures, the “checkpoint signature” and “trafficking of T-cell signature”, were higher in the response subgroup than in the nonresponse subgroup in both the discovery and validation cohorts. Moreover, enrichment of the “checkpoint signature” was correlated with improved overall survival and progression-free survival in both datasets. In addition, we demonstrated that increased baseline levels of three checkpoint molecules (PD-L1, T-cell immunoglobulin mucin receptor 3 and T-cell-specific surface glycoprotein CD28) were common peripheral responsive correlates in both cohorts, thus rendering this “refined checkpoint signature” an ideal candidate for future verification. In the peripheral blood system, the “refined checkpoint signature” may function as a potential biomarker for anti-PD-1/PD-L1 monotherapy in gastrointestinal (GI) cancers.






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this article [and its supplementary information files].
Abbreviations
- anti-PD-1:
-
Anti-programmed cell death 1
- BTLA:
-
B- and T-lymphocyte attenuator
- CNA:
-
Copy number alteration
- CD152:
-
Cluster of differentiation 152
- CR:
-
Complete response
- GEP:
-
Gene expression profiling
- GI:
-
Gastrointestinal
- ICB:
-
Immune checkpoint blockade
- ICI:
-
Immune checkpoint inhibitor
- IF:
-
Immunofluorescence
- IFNs:
-
Interferons; DCs, dendritic cells
- IL18:
-
Interleukin 18
- IL-2:
-
Interleukin-2
- LAG3:
-
Lymphocyte activation gene-3-protein
- mDCs:
-
Mature DCs
- MIF:
-
Migration inhibitory factor
- mIHC:
-
Multiplex immunohistochemistry
- MSI:
-
Microsatellite instability
- ORR:
-
Overall response rate
- OS:
-
Overall survival
- PD:
-
Progressive disease
- PD-L1:
-
Programmed cell death ligand 1
- PFS:
-
Progression-free survival
- PR:
-
Partial response
- RECIST:
-
Response Evaluation Criteria in Solid Tumors
- sCD28:
-
Soluble CD28
- SCLC:
-
Small cell lung cancer
- SD:
-
Standard deviation
- SD:
-
Stable disease
- sPD-L1:
-
Soluble PD-L1
- TGFβ:
-
Transforming growth factor β
- TILs:
-
Tumor infiltrating immune cells
- TIM3:
-
T-cell immunoglobulin mucin receptor 3
- TMB:
-
Tumor mutational burden
- TNFα:
-
Tumor necrosis factor α
References
Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A (2017) Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168(4):707–723. https://doi.org/10.1016/j.cell.2017.01.017
Kim TK, Herbst RS, Chen L (2018) Defining and understanding adaptive resistance in cancer immunotherapy. Trends Immunol 39(8):624–631. https://doi.org/10.1016/j.it.2018.05.001
Havel JJ, Chowell D, Chan TA (2019) The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. https://doi.org/10.1038/s41568-019-0116-x
Gong J, Chehrazi-Raffle A, Reddi S, Salgia R (2018) Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother cancer 6(1):1–18. https://doi.org/10.1186/s40425-018-0316-z
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Diaz LA (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357(6349):409–413
Havel JJ, Chowell D, Chan TA (2019) The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 19(3):133–150
Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X (2018) Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. https://doi.org/10.1126/science.aar3593
Gettinger SN, Choi J, Mani N, Sanmamed MF, Datar I, Sowell R, Du VY, Kaftan E, Goldberg S, Dong W, Zelterman D, Politi K, Kavathas P, Kaech S, Yu X, Zhao H, Schlessinger J, Lifton R, Rimm DL, Chen L, Herbst RS, Schalper KA (2018) A dormant TIL phenotype defines non-small cell lung carcinomas sensitive to immune checkpoint blockers. Nat Commun 9(1):3196. https://doi.org/10.1038/s41467-018-05032-8
Lee S, Margolin K (2011) Cytokines in cancer immunotherapy. Cancers 3(4):3856–3893
Lippitz BE (2013) Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol 14(6):e218-228. https://doi.org/10.1016/s1470-2045(12)70582-x
Hardy-Werbin M, Rocha P, Arpi O, Taus A, Nonell L, Duran X, Villanueva X, Joseph-Pietras D, Nolan L, Danson S, Griffiths R, Lopez-Botet M, Rovira A, Albanell J, Ottensmeier C, Arriola E (2019) Serum cytokine levels as predictive biomarkers of benefit from ipilimumab in small cell lung cancer. Oncoimmunology 8(6):e1593810. https://doi.org/10.1080/2162402x.2019.1593810
Leung AM, Lee AF, Ozao-Choy J, Ramos RI, Hamid O, O’Day SJ, Shin-Sim M, Morton DL, Faries MB, Sieling PA, Lee DJ (2014) Clinical benefit from ipilimumab therapy in melanoma patients may be associated with serum CTLA4 levels. Frontiers in oncology 4:110. https://doi.org/10.3389/fonc.2014.00110
Wang L, Wang H, Chen H, Wang WD, Chen XQ, Geng QR, Xia ZJ, Lu Y (2015) Serum levels of soluble programmed death ligand 1 predict treatment response and progression free survival in multiple myeloma. Oncotarget 6(38):41228–41236. https://doi.org/10.18632/oncotarget.5682
Lu Z, Zou J, Hu Y, Li S, Zhou T, Gong J, Li J, Zhang X, Zhou J, Lu M, Wang X, Peng Z, Qi C, Li Y, Li J, Li Y, Zou J, Du X, Zhang H, Shen L (2019) Serological markers associated with response to immune checkpoint blockade in metastatic gastrointestinal tract cancer. JAMA Netw Open 2(7):e197621. https://doi.org/10.1001/jamanetworkopen.2019.7621
Ozaki K, Leonard WJ (2002) Cytokine and cytokine receptor pleiotropy and redundancy. J Biol Chem 277(33):29355–29358. https://doi.org/10.1074/jbc.R200003200
Chen DS, Mellman I (2013) Oncology meets immunology: the cancer-immunity cycle. Immunity 39(1):1–10. https://doi.org/10.1016/j.immuni.2013.07.012
Berraondo P, Sanmamed MF, Ochoa MC, Etxeberria I, Aznar MA, Perez-Gracia JL, Rodriguez-Ruiz ME, Ponz-Sarvise M, Castanon E, Melero I (2019) Cytokines in clinical cancer immunotherapy. Br J Cancer 120(1):6–15. https://doi.org/10.1038/s41416-018-0328-y
Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, Liu YJ (1999) The nature of the principal type 1 interferon-producing cells in human blood. Science 284(5421):1835–1837. https://doi.org/10.1126/science.284.5421.1835
Guo J, Xiao Y, Iyer R, Lu X, Lake M, Ladror U, Harlan J (2019) Empowering therapeutic antibodies with IFN-alpha for cancer immunotherapy. Plos One 14(8):e0219829. https://doi.org/10.1371/journal.pone.0219829
Gu D, Ao X, Yang Y, Chen Z, Xu X (2018) Soluble immune checkpoints in cancer: production, function and biological significance. J Immunother Cancer 6(1):132. https://doi.org/10.1186/s40425-018-0449-0
Frigola X, Inman BA, Krco CJ, Liu X, Harrington SM, Bulur PA, Dietz AB, Dong H, Kwon ED (2012) Soluble B7–H1: differences in production between dendritic cells and T cells. Immunol Lett 142(1–2):78–82. https://doi.org/10.1016/j.imlet.2011.11.001
Zhou J, Mahoney KM, Giobbie-Hurder A, Zhao F, Lee S, Liao X, Rodig S, Li J, Wu X, Butterfield LH, Piesche M, Manos MP, Eastman LM, Dranoff G, Freeman GJ, Hodi FS (2017) Soluble PD-L1 as a biomarker in malignant melanoma treated with checkpoint blockade. Cancer Immunol Res 5(6):480–492. https://doi.org/10.1158/2326-6066.cir-16-0329
Sun Z, Yi L, Tao H, Huang J, Jin Z, Xiao Y, Feng C, Sun J (2014) Enhancement of soluble CD28 levels in the serum of Graves’ disease. Cent-Eur J Immunol 39(2):216–222. https://doi.org/10.5114/ceji.2014.43726
Goncalves Silva I, Yasinska IM, Sakhnevych SS, Fiedler W, Wellbrock J, Bardelli M, Varani L, Hussain R, Siligardi G, Ceccone G, Berger SM, Ushkaryov YA, Gibbs BF, Fasler-Kan E, Sumbayev VV (2017) The Tim-3-galectin-9 secretory pathway is involved in the immune escape of human acute myeloid leukemia cells. EBioMedicine 22:44–57. https://doi.org/10.1016/j.ebiom.2017.07.018
Funding
This work was supported by grant from the National Key Sci-Tech Special Project of China (No. 2018ZX10302207).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Internal Review and the Committee of the Fifth Medical Center, General Hospital of the PLA, Ethics Committee of Zhongshan Hospital Affiliated to Fudan University and was performed in accordance with the Declaration of HELSINKI.
Consent to participate
Informed consent was obtained from each patient before sample collection.
Consent for publication
We declare that this manuscript is original, has not been published before, and is not currently under consideration for publication elsewhere.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, C., Wu, L., Liang, D. et al. Identification of immune checkpoint and cytokine signatures associated with the response to immune checkpoint blockade in gastrointestinal cancers. Cancer Immunol Immunother 70, 2669–2679 (2021). https://doi.org/10.1007/s00262-021-02878-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-021-02878-8