Abstract
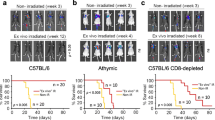
Maml1 is emerging as a coactivator of many signaling pathways, including the Notch and Wnt pathways. Targeting Maml1 in melanoma cells efficiently knocks down the downstream transcriptional repressors Hey1 and Hes1, resulting in melanoma cell senescence, cellular differentiation, and increased melanin production. Significantly, higher IFNβ and chemokine gene transcripts have been observed, together with increased STAT1 and decreased STAT3 and NF-κB signaling activities. Although decreased cell proliferation contributes to slower tumor growth in vivo, the depletion of NK and CD8+ T cells in an shMaml1-B16 tumor carrier mouse leads to more rapid tumor growth than that observed in control shC002-B16 tumors. This result demonstrates that the knockdown of Maml1 transcription and function contributes to increased immune surveillance. The knockdown of Maml1 transcription in the human melanoma cell line M537 also results in senescence, IFNβ upregulation, increased chemokine gene expression, and greater NK and CD8+ T cell migration in a transwell system. This study demonstrated that targeting Maml1-induced tumor cell senescence and differentiation may alter the tumor microenvironment and cytokine and chemokine profiles and may also promote innate and adaptive immune cell infiltration and function.




Similar content being viewed by others
Abbreviations
- Maml1 :
-
Mastermind-like protein-1
- siMaml1 :
-
Small interfering Maml1 RNA
- Hes1:
-
Hairy/enhancer-of-split 1
- Hey1:
-
Hairy/enhancer-of-split related with YRPW motif 1
- CCL2:
-
MCP-1, monocyte chemoattractant-1
- CCL3:
-
Macrophage inflammatory protein-1α (MIP-1α)
- CCL5:
-
Regulated upon activation normal T cell expressed and secreted (RANTES)
- CCL18:
-
Macrophage inflammatory protein-4 (MIP-4, PARC)
- CXCL9:
-
Monokine induced by gamma interferon (Mig)
- CXCL10:
-
Interferon gamma-induced protein-10 (IP-10)
- CXCL11:
-
Interferon-inducible T cell alpha chemoattractant (I-TAC, IP-9)
References
Takebe N, Harris PJ, Warren RQ, Ivy SP (2011) Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol 8:97–106
Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB (2009) Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov 8:806–823
Crea F, Duhagon MA, Farrar WL, Danesi R (2011) Pharmacogenomics and cancer stem cells: a changing landscape? Trends Pharmacol Sci 32:487–494
Radtke F, Raj K (2003) The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer 3:756–767
Hansson EM, Lendahl U, Chapman G (2004) Notch signaling in development and disease. Semin Cancer Biol 14:320–328
Nickoloff BJ, Osborne BA, Miele L (2003) Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene 22:6598–6608
Kang S, Yang C, Luo R (2008) Induction of CCL2 by siMAML1 through upregulation of TweakR in melanoma cells. Biochem Biophys Res Commun 372:629–633
Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233
Alves-Guerra MC, Ronchini C, Capobianco AJ (2007) Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res 67:8690–8698
Shimizu T, Kagawa T, Inoue T, Nonaka A, Takada S, Aburatani H, Taga T (2008) Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol 28:7427–7441
Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ (2008) Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene 27:1489–1500
Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, Wallace VA (2009) Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol 184:101–112
Hatton BA, Villavicencio EH, Pritchard J, LeBlanc M, Hansen S, Ulrich M, Ditzler S, Pullar B, Stroud MR, Olson JM (2010) Notch signaling is not essential in sonic hedgehog-activated medulloblastoma. Oncogene 29:3865–3872
Sang L, Roberts JM, Coller HA (2010) Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med 16:17–26
Schreck KC, Taylor P, Marchionni L, Gopalakrishnan V, Bar EE, Gaiano N, Eberhart CG (2010) The Notch target Hes1 directly modulates Gli1 expression and Hedgehog signaling: a potential mechanism of therapeutic resistance. Clin Cancer Res 16:6060–6070
Sang L, Coller HA (2009) Fear of commitment: Hes1 protects quiescent fibroblasts from irreversible cellular fates. Cell Cycle 8:2161–2167
Sang L, Coller HA, Roberts JM (2008) Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321:1095–1100
Wu L, Kobayashi K, Sun T, Gao P, Liu J, Nakamura M, Weisberg E, Mukhopadhyay NK, Griffin JD (2004) Cloning and functional characterization of the murine mastermind-like 1 (Maml1) gene. Gene 328:153–165
Jin B, Shen H, Lin S, Li JL, Chen Z, Griffin JD, Wu L (2010) The mastermind-like 1 (MAML1) co-activator regulates constitutive NF-kappaB signaling and cell survival. J Biol Chem 285:14356–14365
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE (2009) Direct inhibition of the NOTCH transcription factor complex. Nature 462:182–188
Bartlett DW, Davis ME (2006) Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res 34:322–333
Kang S, Xie J, Ma S, Liao W, Zhang J, Luo R (2010) Targeted knock down of CCL22 and CCL17 by siRNA during DC differentiation and maturation affects the recruitment of T subsets. Immunobiology 215:153–162
Hiroi M, Ohmori Y (2003) Constitutive nuclear factor kappaB activity is required to elicit interferon-gamma-induced expression of chemokine CXC ligand 9 (CXCL9) and CXCL10 in human tumour cell lines. Biochem J 376:393–402
Biswas SK, Gangi L, Paul S, Schioppa T, Saccani A, Sironi M, Bottazzi B, Doni A, Vincenzo B, Pasqualini F, Vago L, Nebuloni M, Mantovani A, Sica A (2006) A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation). Blood 107:2112–2122
Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F (2008) Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep 9:377–383
Kavanagh E, Joseph B (2011) The hallmarks of CDKN1C (p57, KIP2) in cancer. Biochim Biophys Acta 1816:50–56
Jin B, Shen H, Lin S, Li JL, Chen Z, Griffin JD, Wu L (2010) The mastermind-like 1 (MAML1) co-activator regulates constitutive NF-kappaB signaling and cell survival. J Biol Chem 285:14356–14365
Cao Q, Kaur C, Wu CY, Lu J, Ling EA (2011) Nuclear factor-kappa beta regulates Notch signaling in production of proinflammatory cytokines and nitric oxide in murine BV-2 microglial cells. Neuroscience 192:140–154
Bhoopathi P, Chetty C, Dontula R, Gujrati M, Dinh DH, Rao JS, Lakka SS (2011) SPARC stimulates neuronal differentiation of medulloblastoma cells via the Notch1/STAT3 pathway. Cancer Res 71:4908–4919
Lee JH, Suk J, Park J, Kim SB, Kwak SS, Kim JW, Lee CH, Byun B, Ahn JK, Joe CO (2009) Notch signal activates hypoxia pathway through HES1-dependent SRC/signal transducers and activators of transcription 3 pathway. Mol Cancer Res 7:1663–1671
Ma J, Meng Y, Kwiatkowski DJ, Chen X, Peng H, Sun Q, Zha X, Wang F, Wang Y, Jing Y, Zhang S, Chen R, Wang L, Wu E, Cai G, Malinowska-Kolodziej I, Liao Q, Liu Y, Zhao Y, Xu K, Dai J, Han J, Wu L, Zhao RC, Shen H, Zhang H (2010) Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Invest 120:103–114
Lee H, Deng J, Xin H, Liu Y, Pardoll D, Yu H (2011) A requirement of STAT3 DNA binding precludes Th-1 immunostimulatory gene expression by NF-kappaB in tumors. Cancer Res 71:3772–3780
Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H (2009) Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 15:283–293
Lee H, Pal SK, Reckamp K, Figlin RA, Yu H (2011) STAT3: a target to enhance antitumor immune response. Curr Top Microbiol Immunol 344:41–59
Yu H, Pardoll D, Jove R (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9:798–809
Regis G, Pensa S, Boselli D, Novelli F, Poli V (2008) Ups and downs: the STAT1:STAT3 seesaw of Interferon and gp130 receptor signalling. Semin Cell Dev Biol 19:351–359
Pinho AV, Rooman I, Reichert M, De Medts N, Bouwens L, Rustgi AK, Real FX (2011) Adult pancreatic acinar cells dedifferentiate to an embryonic progenitor phenotype with concomitant activation of a senescence programme that is present in chronic pancreatitis. Gut 60:958–966
Lleonart ME, Artero-Castro A, Kondoh H (2009) Senescence induction; a possible cancer therapy. Mol Cancer 8:3
Roninson IB (2003) Tumor cell senescence in cancer treatment. Cancer Res 63:2705–2715
Schmitt CA (2007) Cellular senescence and cancer treatment. Biochim Biophys Acta 1775:5–20
Kahlem P, Dorken B, Schmitt CA (2004) Cellular senescence in cancer treatment: friend or foe? J Clin Invest 113:169–174
Freund A, Orjalo AV, Desprez PY, Campisi J (2010) Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med 16:238–246
Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW (2007) Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445:656–660
Rakhra K, Bachireddy P, Zabuawala T, Zeiser R, Xu L, Kopelman A, Fan AC, Yang Q, Braunstein L, Crosby E, Ryeom S, Felsher DW (2010) CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 18:485–498
Acknowledgments
This work was supported by the Province of Guangdong Science Grant (10251051501000008).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kang, S., Xie, J., Miao, J. et al. A knockdown of Maml1 that results in melanoma cell senescence promotes an innate and adaptive immune response. Cancer Immunol Immunother 62, 183–190 (2013). https://doi.org/10.1007/s00262-012-1318-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-012-1318-1