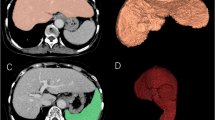
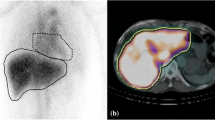
Abstract
Purpose
To assess the utility of a machine-learning approach for predicting liver function based on technetium-99 m-galactosyl serum albumin (99mTc-GSA) single photon emission computed tomography (SPECT)/CT.
Methods
One hundred twenty-eight patients underwent a 99mTc-GSA SPECT/CT-based liver function evaluation. All were classified into the low liver-damage or high liver-damage group. Four clinical (age, sex, background liver disease and histological type) and 8 quantitative 99mTc-GSA SPECT/CT features (receptor index [LHL15], clearance index [HH15], liver-SUVmax, liver-SUVmean, heart-SUVmax, metabolic volume of liver [MVL], total lesion GSA [TL-GSA, liver-SUVmean × MVL] and SUVmax ratio [liver-SUVmax/heart-SUVmax]) were obtained. To predict high liver damage, a machine learning classification with features selection based on Gini impurity and principal component analysis (PCA) were performed using a support vector machine and a random forest (RF) with a five-fold cross-validation scheme. To overcome imbalanced data, stratified sampling was used. The ability to predict high liver damage was evaluated using a receiver operating characteristic (ROC) curve analysis.
Results
Four indices (LHL15, HH15, heart SUVmax and SUVmax ratio) yielded high areas under the ROC curves (AUCs) for predicting high liver damage (range: 0.89–0.93). In a machine learning classification, the RF with selected features (heart SUVmax, SUVmax ratio, LHL15, HH15, and background liver disease) and PCA model yielded the best performance for predicting high liver damage (AUC = 0.956, sensitivity = 96.3%, specificity = 90.0%, accuracy = 91.4%).
Conclusion
A machine-learning approach based on clinical and quantitative 99mTc-GSA SPECT/CT parameters might be useful for predicting liver function.


Similar content being viewed by others
References
Hoekstra LT, de Graaf W, Nibourg GA, et al (2013) Physiological and biochemical basis of clinical liver function tests: a review. Ann Surg 257:27-36
Ashwell G, Morell AG (1974) The role of surface carbohydrates in the hepatic recognition and transport of circulating glycoproteins. Adv Enzymol Relat Areas Mol Biol 41: 99-128
Sobue G, Kosaka A (1980) Asialoglycoproteinemia in a case of primary hepatic cancer. Hepatogastroenterology 27:200-203.
Marshall JS, Green AM, Pensky J, Williams S, Zinn A, Carlson DM (1974) Measurement of circulating desialylated glycoproteins and correlation with hepatocellular damage. J Clin Invest 54:555-562.
Sawamura T, Nakada H, Hazama H, Shiozaki Y, Sameshima Y, Tashiro Y (1984) Hyperasialoglycoproteinemia in patients with chronic liver diseases and/or liver cell carcinoma. Asialoglycoprotein receptor in cirrhosis and liver cell carcinoma. Gastroenterology 87:1217-1221
Okabayashi T, Shima Y, Morita S, et al (2017) Liver function assessment using technetium 99m-Galactosyl single-photon emission computed tomography/CT fusion imaging: a prospective trial. J Am Coll Surg 225:789-797
Yoshida M, Beppu T, Shiraishi S, et al (2015) (99m)Tc-GSA SPECT/CT fused images for assessment of hepatic function and hepatectomy planning. Ann Transl Med 3:17
Sasaki N, Shiomi S, Iwata Y, et al (1999) Clinical usefulness of scintigraphy with 99mTc-galactosyl-human serum albumin for prognosis of cirrhosis of the liver. J Nucl Med 40:1652-1656
Shiomi S, Sasaki N, Tamori A, et al (1999) Use of scintigraphy with 99mtechnetium galactosyl human serum albumin for staging of primary biliary cirrhosis and assessment of prognosis. J Gastroenterol Hepatol 14:566-571
Buck AK, Nekolla S, Ziegler S, et al (2008) SPECT/CT. J Nucl Med 49:1305-1319.
Kaneta T, Ogawa M, Daisaki H, Nawata S, Yoshida K, Inoue T (2016) SUV measurement of normal vertebrae using SPECT/CT with Tc-99 m methylene diphosphonate. Am J Nucl Med Mol Imaging 6:262-268
Suh MS, Lee WW, Kim YK, Yun PY, Kim SE (2016) Maximum standardized uptake value of (99 m)Tc hydroxymethylene diphosphonate SPECT/CT for the evaluation of temporomandibular joint disorder. Radiology 280:890-896
Beck M, Sanders JC, Ritt P, Reinfelder J, Kuwert T (2016) Longitudinal analysis of bone metabolism using SPECT/CT and (99 m)Tc-diphosphono-propanedicarboxylic acid: comparison of visual and quantitative analysis. EJNMMI Res 6:60
Tokorodani R, Sumiyoshi T, Okabayashi T, et al (2019) Liver fibrosis assessment using 99 mTc-GSA SPECT/CT fusion imaging. Jpn J Radiol 37:315-320
Erickson BJ, Korfiatis P, Akkus Z, Kline TL (2017) Machine learning for medical imaging. Radiographics 37:505–515
Waljee AK, Higgins PD (2010) Machine learning in medicine: a primer for physicians. Am J Gastroenterol 105:1224–1246
Gao X, Chu C, Li Y, et al (2015) The method and efficacy of support vector machine classifiers based on texture features and multi-resolution histogram from (18)F-FDG PET-CT images for the evaluation of mediastinal lymph nodes in patients with lung cancer. Eur J Radiol 84:312-317
Ypsilantis PP, Siddique M, Sohn HM, et al (2015) Predicting Response to Neoadjuvant Chemotherapy with PET Imaging Using Convolutional Neural Networks. PLoS One 10:e0137036
Ahn HK, Lee H, Kim SG, Hyun SH (2019) Pre-treatment 18F-FDG PET-based radiomics predict survival in resected non-small cell lung cancer. Clin Radiol 74:467-473
Nakajima K, Kinuya K, Mizutani Y, et al (1999) Simple scintigraphic parameters with Tc-99m galactosyl human serum albumin for clinical staging of chronic hepatocellular dysfunction. Ann Nucl Med 13:5-11
Ikai I, Arii S, Kojiro M, et al (2004) Reevaluation of prognostic factors for survival after liver resection in patients with hepatocellular carcinoma in a Japanese nationwide survey. Cancer 101:796-802
Sian H, Purnami SW (2015) Combine sampling support vector machine for imbalanced data classification. Procedia Computer Science 72:59-66
Xie Y, Jiang B, Gong E, et al (2019) Use of gradient boosting machine learning to predict patient outcome in acute ischemic stroke on the basis of imaging, demographic, and clinical information. AJR Am J Roentgenol 212:44-51
Jung Y (2018) Multiple predicting K-fold cross-validation for model selection. J Nonparametr Stat 30:197-215
Cook JA, Ranstam J (2016) Overfitting. Br J Surg 103:1814
Breiman L (2001) Random forests. Mach Learn 45:5-32
Rahman R, Kodesh A, Levine SZ, Sandin S, Reichenberg A, Schlessinger A (2020). Identification of newborns at risk for autism using electronic medical records and machine learning. Eur Psychiatry 63:e22
Hotta M, Minamimoto R, Miwa K (2019) 11C-methionine-PET for differentiating recurrent brain tumor from radiation necrosis: radiomics approach with random forest classifier. Sci Rep 9:15666.
Demsar J, Curk T, Erjavec A, et al (2013) Orange: data mining toolbox in Python. J Machine Learn Res 14:2349-2353
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837-845
Kitajima K, Futani H, Fujiwara M, et al (2018) Usefulness of quantitative bone single photon emission computed tomography/computed tomography for evaluating response to neoadjuvant chemotherapy in a patient with periosteal osteosarcoma. Cureus 10:e3655
Zeintl J, Vija AH, Yahil A, Hornegger J, Kuwert T (2010) Quantitative accuracy of clinical 99 mTc SPECT/CT using ordered-subset expectation maximization with 3-dimensional resolution recovery, attenuation, and scatter correction. J Nucl Med 51:921-928
Bailey DL, Willowson KP (2013) An evidence-based review of quantitative SPECT imaging and potential clinical applications. J Nucl Med 54:83-89
Asl BM, Setarehdan SK, Mohebbi M (2008) Support vector machine-based arrhythmia classification using reduced features of heart rate variability signal. Artif Intell Med 44:51-64
Sakai A, Onishi Y, Matsui M, et al (2020) A method for the automated classification of benign and malignant masses on digital breast tomosynthesis images using machine learning and radiomic features. Radiol Phys Technol 13:27-36
Rajula HSR, Verlato G, Manchia M, Antonucci N, Fanos V (2020) Comparison of conventional statistical methods with machine learning in medicine: diagnosis, drug development, and treatment. Medicina (Kaunas) 56:455
Ngiam, KY Khor, IW (2019) Big data and machine learning algorithms for health-care delivery. Lancet Oncol 20: e262–e273
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was waived by the institutional review board for this retrospective study.
Research involving human and animal rights
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nakajo, M., Jinguji, M., Tani, A. et al. Application of a machine learning approach to characterization of liver function using 99mTc-GSA SPECT/CT. Abdom Radiol 46, 3184–3192 (2021). https://doi.org/10.1007/s00261-021-02985-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-021-02985-1