Abstract
Purpose
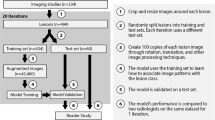
To develop a deep convolutional neural network (CNN) model to categorize multiphase CT and MRI liver observations using the liver imaging reporting and data system (LI-RADS) (version 2014).
Methods
A pre-existing dataset comprising 314 hepatic observations (163 CT, 151 MRI) with corresponding diameters and LI-RADS categories (LR-1–5) assigned in consensus by two LI-RADS steering committee members was used to develop two CNNs: pre-trained network with an input of triple-phase images (training with transfer learning) and custom-made network with an input of quadruple-phase images (training from scratch). The dataset was randomly split into training, validation, and internal test sets (70:15:15 split). The overall accuracy and area under receiver operating characteristic curve (AUROC) were assessed for categorizing LR-1/2, LR-3, LR-4, and LR-5. External validation was performed for the model with the better performance on the internal test set using two external datasets (EXT-CT and EXT-MR: 68 and 44 observations, respectively).
Results
The transfer learning model outperformed the custom-made model: overall accuracy of 60.4% and AUROCs of 0.85, 0.90, 0.63, 0.82 for LR-1/2, LR-3, LR-4, LR-5, respectively. On EXT-CT, the model had an overall accuracy of 41.2% and AUROCs of 0.70, 0.66, 0.60, 0.76 for LR-1/2, LR-3, LR-4, LR-5, respectively. On EXT-MR, the model had an overall accuracy of 47.7% and AUROCs of 0.88, 0.74, 0.69, 0.79 for LR-1/2, LR-3, LR-4, LR-5, respectively.
Conclusion
Our study shows the feasibility of CNN for assigning LI-RADS categories from a relatively small dataset but highlights the challenges of model development and validation.






Similar content being viewed by others
Abbreviations
- LI-RADS:
-
Liver imaging reporting and data system
- HCC:
-
Hepatocellular carcinoma
- CNN:
-
Convolutional neural network
- ROI:
-
Region of interest
- AUROC:
-
Area under receiver operating characteristic curve
References
American College of Radiology ACR LI-RADS v2014. https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS/LI-RADS-v2014. Accessed December 11, 2018.
Fowler KJ, Tang A, Santillan C, Bhargavan-Chatfield M, Heiken J, Jha RC, Weinreb J, Hussain H, Mitchell DG, Bashir MR, Costa EAC, Cunha GM, Coombs L, Wolfson T, Gamst AC, Brancatelli G, Yeh B, Sirlin CB (2018) Interreader Reliability of LI-RADS Version 2014 Algorithm and Imaging Features for Diagnosis of Hepatocellular Carcinoma: A Large International Multireader Study. Radiology 286 (1):173-185. https://doi.org/10.1148/radiol.2017170376
Schellhaas B, Hammon M, Strobel D, Pfeifer L, Kielisch C, Goertz RS, Cavallaro A, Janka R, Neurath MF, Uder M, Seuss H (2018) Interobserver and intermodality agreement of standardized algorithms for non-invasive diagnosis of hepatocellular carcinoma in high-risk patients: CEUS-LI-RADS versus MRI-LI-RADS. European radiology 28 (10):4254-4264. https://doi.org/10.1007/s00330-018-5379-1
Barth BK, Donati OF, Fischer MA, Ulbrich EJ, Karlo CA, Becker A, Seifert B, Reiner CS (2016) Reliability, Validity, and Reader Acceptance of LI-RADS-An In-depth Analysis. Academic radiology 23 (9):1145-1153. https://doi.org/10.1016/j.acra.2016.03.014
Davenport MS, Khalatbari S, Liu PS, Maturen KE, Kaza RK, Wasnik AP, Al-Hawary MM, Glazer DI, Stein EB, Patel J, Somashekar DK, Viglianti BL, Hussain HK (2014) Repeatability of diagnostic features and scoring systems for hepatocellular carcinoma by using MR imaging. Radiology 272 (1):132-142. https://doi.org/10.1148/radiol.14131963
Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR (2016) Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. Jama 316 (22):2402-2410. https://doi.org/10.1001/jama.2016.17216
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542 (7639):115-118. https://doi.org/10.1038/nature21056
Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak J, Hermsen M, Manson QF, Balkenhol M, Geessink O, Stathonikos N, van Dijk MC, Bult P, Beca F, Beck AH, Wang D, Khosla A, Gargeya R, Irshad H, Zhong A, Dou Q, Li Q, Chen H, Lin HJ, Heng PA, Hass C, Bruni E, Wong Q, Halici U, Oner MU, Cetin-Atalay R, Berseth M, Khvatkov V, Vylegzhanin A, Kraus O, Shaban M, Rajpoot N, Awan R, Sirinukunwattana K, Qaiser T, Tsang YW, Tellez D, Annuscheit J, Hufnagl P, Valkonen M, Kartasalo K, Latonen L, Ruusuvuori P, Liimatainen K, Albarqouni S, Mungal B, George A, Demirci S, Navab N, Watanabe S, Seno S, Takenaka Y, Matsuda H, Ahmady Phoulady H, Kovalev V, Kalinovsky A, Liauchuk V, Bueno G, Fernandez-Carrobles MM, Serrano I, Deniz O, Racoceanu D, Venancio R (2017) Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. Jama 318 (22):2199-2210. https://doi.org/10.1001/jama.2017.14585
Chilamkurthy S, Ghosh R, Tanamala S, Biviji M, Campeau NG, Venugopal VK, Mahajan V, Rao P, Warier P (2018) Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet (London, England) 392 (10162):2388-2396. https://doi.org/10.1016/s0140-6736(18)31645-3
De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, Askham H, Glorot X, O’Donoghue B, Visentin D, van den Driessche G, Lakshminarayanan B, Meyer C, Mackinder F, Bouton S, Ayoub K, Chopra R, King D, Karthikesalingam A, Hughes CO, Raine R, Hughes J, Sim DA, Egan C, Tufail A, Montgomery H, Hassabis D, Rees G, Back T, Khaw PT, Suleyman M, Cornebise J, Keane PA, Ronneberger O (2018) Clinically applicable deep learning for diagnosis and referral in retinal disease. Nature medicine 24 (9):1342-1350. https://doi.org/10.1038/s41591-018-0107-6
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv. https://arxiv.org/abs/1409.1556. vol 1409.1556.
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L (2015) ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision 115 (3):211-252. https://doi.org/10.1007/s11263-015-0816-y
Yamashita R, Nishio M, Do RKG, Togashi K (2018) Convolutional neural networks: an overview and application in radiology. Insights into imaging 9 (4):611-629. https://doi.org/10.1007/s13244-018-0639-9
Kluyver T, Ragan-Kelley B, Pérez F, Granger BE, Bussonnier M, Frederic J, Kelley K, Hamrick JB, Grout J, Corlay S (2016) Jupyter Notebooks-a publishing format for reproducible computational workflows. In: Loizides F, Scmidt B (eds) Positioning and Power in Academic Publishing: Players, Agents and Agendas. IOS Press, pp 87-90. https://doi.org/10.3233/978-1-61499-649-1-87
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33 (1):159-174. https://doi.org/10.2307/2529310
Jones E, Oliphant T, Peterson P, et al SciPy: open source scientific tools for Python, 2001-, http://www.scipy.org/. Accessed on December 10, 2018.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V (2011) Scikit-learn: Machine learning in Python. Journal of machine learning research 12:2825-2830. doi:Not available
R Development Core Team (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology 286 (3):887-896. https://doi.org/10.1148/radiol.2017170706
Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK (2018) Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS medicine 15 (11):e1002683. https://doi.org/10.1371/journal.pmed.1002683
Park SH (2019) Diagnostic Case-Control versus Diagnostic Cohort Studies for Clinical Validation of Artificial Intelligence Algorithm Performance. Radiology 290 (1):272-273. https://doi.org/10.1148/radiol.2018182294
Jha RC, Mitchell DG, Weinreb JC, Santillan CS, Yeh BM, Francois R, Sirlin CB (2014) LI-RADS categorization of benign and likely benign findings in patients at risk of hepatocellular carcinoma: a pictorial atlas. AJR American journal of roentgenology 203 (1):W48-69. https://doi.org/10.2214/ajr.13.12169
Acknowledgements
We thank Joanne Chin for editorial assistance.
Funding
Supported by JSPS Overseas Research Fellowships (R.Y.) (Japan Society for the Promotion of Science (JSPS/OT/290125)) and the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748 (R.Y. and R.K.G.D.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent
For this type of study, formal consent is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yamashita, R., Mittendorf, A., Zhu, Z. et al. Deep convolutional neural network applied to the liver imaging reporting and data system (LI-RADS) version 2014 category classification: a pilot study. Abdom Radiol 45, 24–35 (2020). https://doi.org/10.1007/s00261-019-02306-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-019-02306-7