Abstract
Purpose
Multiple myeloma (MM) is a malignant disease characterized by the secretion of monoclonal immunoglobulins and has a high demand for amino acids. [11C]methionine total-body PET is capable of noninvasive dynamic monitoring of radiotracer in vivo, thus providing a way to reveal the dynamic changes of myeloma metabolism. This study aims to analyze the metabolic process of [11C]methionine based on kinetic modeling, and to preliminary reveal its application value in MM.
Methods
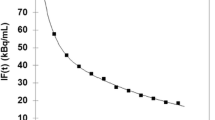
Dynamic total-body [11C]methionine PET/CT was conducted with uEXPLORER in 12 subjects (9 MM patients and 3 controls). The tissue time activity curves (TACs) of organs and bone marrows were extracted. Model fitting of TACs was operated using PMOD Kinetic Modeling. After validation by Goodness of fit (GOF), the reversible two-tissue compartment model (2T4k) was used to further analysis. R software was used to analyze the correlation between kinetic parameters and clinical indicators.
Results
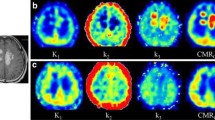
The 2T4k has passed the criterion of GOF and was used to fit the data of 0-20 minutes. The [11C]methionine net uptake rate (Ki) was significantly higher in the MM lesions than in the non-myeloma controls (control: 0.040±0.007 mL/g/min, MM: 0.171±0.108 mL/g/min, p=0.009). The Ki values were found to be correlated with M protein levels in MM patients. MM patients with t(4;14) translocations had an elevated k4 value compared with t(4;14) negative patients.
Conclusion
MM lesions have a propensity for uptake of [11C]methionine. The serum levels of M protein are correlated with [11C]methionine uptake rate in myeloma. Metabolic classification based on the k4 value may be a promising strategy for risk stratification in MM.






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Shapiro YN, Peppercorn JM, Yee AJ, Branagan AR, Raje NS, Donnell EKO. Lifestyle considerations in multiple myeloma. Blood Cancer J. 2021;11:172. https://doi.org/10.1038/s41408-021-00560-x.
Hammerton K, Cooper DA, Duckett M, Penny R. Biosynthesis of immunoglobulin in human immunoproliferative diseases. I. Kinetics of synthesis and secretion of immunoglobulin and protein by bone marrow cells in myeloma. J Immunol. 1978;121:409–17.
Khor HK, Jacoby ME, Squier TC, Chu GC, Chelius D. Identification of methionine sulfoxide diastereomers in immunoglobulin gamma antibodies using methionine sulfoxide reductase enzymes. mAbs. 2010;2:299–308. https://doi.org/10.4161/mabs.2.3.11755.
Kim D, Chun JH, Kim SH, Moon JH, Kang SG, Chang JH, et al. Re-evaluation of the diagnostic performance of (11)C-methionine PET/CT according to the 2016 WHO classification of cerebral gliomas. Eur J Nucl Med Mol Imaging. 2019;46:1678–84. https://doi.org/10.1007/s00259-019-04337-0.
Morales-Lozano MI, Viering O, Samnick S, Rodriguez-Otero P, Buck AK, Marcos-Jubilar M, et al. (18)F-FDG and (11)C-Methionine PET/CT in Newly Diagnosed Multiple Myeloma Patients: Comparison of Volume-Based PET Biomarkers. Cancers. 2020:12. https://doi.org/10.3390/cancers12041042.
Matteucci F, Paganelli G, Martinelli G, Cerchione C. PET/CT in Multiple Myeloma: Beyond FDG. Front Oncol. 2020;10:622501. https://doi.org/10.3389/fonc.2020.622501.
Wang G, Nardo L, Parikh M, Abdelhafez YG, Li E, Spencer BA, et al. Total-Body PET Multiparametric Imaging of Cancer Using a Voxel-wise Strategy of Compartmental Modeling. J Nucl Med. 2021;63(8):1274–81. https://doi.org/10.2967/jnumed.121.262668.
Wu Y, Feng T, Zhao Y, Xu T, Fu F, Huang Z, et al. Whole-body Parametric Imaging of FDG PET using uEXPLORER with Reduced Scan Time. J Nucl Med. 2021;63(4):622–8. https://doi.org/10.2967/jnumed.120.261651.
Mitterhauser M, Wadsak W, Krcal A, Schmaljohann J, Eidherr H, Schmid A, et al. New aspects on the preparation of [11C]Methionine--a simple and fast online approach without preparative HPLC. Appl Radiat Isot. 2005;62:441–5. https://doi.org/10.1016/j.apradiso.2004.07.006.
Zhang X, Xie Z, Berg E, Judenhofer MS, Liu W, Xu T, et al. Total-Body Dynamic Reconstruction and Parametric Imaging on the uEXPLORER. J Nucl Med. 2020;61:285–91. https://doi.org/10.2967/jnumed.119.230565.
Lucignani G, Paganelli G, Bombardieri E. The use of standardized uptake values for assessing FDG uptake with PET in oncology: a clinical perspective. Nucl Med Commun. 2004;25:651–6. https://doi.org/10.1097/01.mnm.0000134329.30912.49.
Bolcaen J, Descamps B, Deblaere K, Boterberg T, De Vos PF, Kalala JP, et al. (18)F-fluoromethylcholine (FCho), (18)F-fluoroethyltyrosine (FET), and (18)F-fluorodeoxyglucose (FDG) for the discrimination between high-grade glioma and radiation necrosis in rats: a PET study. Nucl Med Biol. 2015;42:38–45. https://doi.org/10.1016/j.nucmedbio.2014.07.006.
Berghmans T, Dusart M, Paesmans M, Hossein-Foucher C, Buvat I, Castaigne C, et al. Primary tumor standardized uptake value (SUVmax) measured on fluorodeoxyglucose positron emission tomography (FDG-PET) is of prognostic value for survival in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis (MA) by the European Lung Cancer Working Party for the IASLC Lung Cancer Staging Project. J Thorac Oncol. 2008;3:6–12. https://doi.org/10.1097/JTO.0b013e31815e6d6b.
Kinahan PE, Fletcher JW. Positron emission tomography-computed tomography standardized uptake values in clinical practice and assessing response to therapy. Semin Ultrasound CT MR. 2010;31:496–505. https://doi.org/10.1053/j.sult.2010.10.001.
Jodal L, Jensen SB, Nielsen OL, Afzelius P, Borghammer P, Alstrup AKO, et al. Kinetic Modelling of Infection Tracers [(18)F]FDG, [(68)Ga]Ga-Citrate, [(11)C]Methionine, and [(11)C]Donepezil in a Porcine Osteomyelitis Model. Contrast Media Mol Imaging. 2017;2017:9256858. https://doi.org/10.1155/2017/9256858.
Grkovski M, Goel R, Krebs S, Staton KD, Harding JJ, Mellinghoff IK, et al. Pharmacokinetic Assessment of (18)F-(2S,4R)-4-Fluoroglutamine in Patients with Cancer. J Nucl Med. 2020;61:357–66. https://doi.org/10.2967/jnumed.119.229740.
Polvoy I, Seo Y, Parker M, Stewart M, Siddiqua K, Manacsa HS, et al. Imaging joint infections using D-methyl-(11)C-methionine PET/MRI: initial experience in humans. Eur J Nucl Med Mol Imaging. 2022;49:3761–71. https://doi.org/10.1007/s00259-022-05858-x.
Nakajo M, Ojima S, Kawakami H, Tani A, Hirayama A, Jinguji M, et al. Value of Patlak Ki images from (18)F-FDG-PET/CT for evaluation of the relationships between disease activity and clinical events in cardiac sarcoidosis. Sci Rep. 2021;11:2729. https://doi.org/10.1038/s41598-021-82217-0.
Lubberink M, Eriksson O. [(11)C]5-Hydroxy-tryptophan model for quantitative assessment of in vivo serotonin biosynthesis, retention and degradation in the endocrine pancreas. Am J Nucl Med Mol Imaging. 2020;10:226–34.
Kletting P, Schimmel S, Kestler HA, Hanscheid H, Luster M, Fernandez M, et al. Molecular radiotherapy: the NUKFIT software for calculating the time-integrated activity coefficient. Med Phys. 2013;40:102504. https://doi.org/10.1118/1.4820367.
Hardiansyah D, Riana A, Kletting P, Zaid NRR, Eiber M, Pawiro SA, et al. A population-based method to determine the time-integrated activity in molecular radiotherapy. EJNMMI Phys. 2021;8:82. https://doi.org/10.1186/s40658-021-00427-x.
Terpos E. Biochemical markers of bone metabolism in multiple myeloma. Cancer Treat Rev. 2006;32(Suppl 1):15–9. https://doi.org/10.1016/s0305-7372(06)80004-6.
Dankerl A, Liebisch P, Glatting G, Friesen C, Blumstein NM, Kocot D, et al. Multiple Myeloma: Molecular Imaging with 11C-Methionine PET/CT--Initial Experience. Radiology. 2007;242:498–508. https://doi.org/10.1148/radiol.2422051980.
Schimmelpennink MC, Vorselaars ADM, Veltkamp M, Keijsers RGM. Quantification of pulmonary disease activity in sarcoidosis measured with (18)F-FDG PET/CT: SUVmax versus total lung glycolysis. EJNMMI Res. 2019;9:54. https://doi.org/10.1186/s13550-019-0505-x.
Bolzoni M, Chiu M, Accardi F, Vescovini R, Airoldi I, Storti P, et al. Dependence on glutamine uptake and glutamine addiction characterize myeloma cells: a new attractive target. Blood. 2016;128:667–79. https://doi.org/10.1182/blood-2016-01-690743.
Lapa C, Garcia-Velloso MJ, Luckerath K, Samnick S, Schreder M, Otero PR, et al. (11)C-Methionine-PET in Multiple Myeloma: A Combined Study from Two Different Institutions. Theranostics. 2017;7:2956–64. https://doi.org/10.7150/thno.20491.
Hsu H, Yu YM, Babich JW, Burke JF, Livni E, Tompkins RG, et al. Measurement of muscle protein synthesis by positron emission tomography with L-[methyl-11C]methionine. Proc Natl Acad Sci U S A. 1996;93:1841–6. https://doi.org/10.1073/pnas.93.5.1841.
Fischman AJ, Yu YM, Livni E, Babich JW, Young VR, Alpert NM, et al. Muscle protein synthesis by positron-emission tomography with L-[methyl-11C]methionine in adult humans. Proc Natl Acad Sci U S A. 1998;95:12793–8. https://doi.org/10.1073/pnas.95.22.12793.
Martinov MV, Vitvitsky VM, Banerjee R, Ataullakhanov FI. The logic of the hepatic methionine metabolic cycle. Biochim Biophys Acta. 2010;1804:89–96. https://doi.org/10.1016/j.bbapap.2009.10.004.
Mu L, Kramer SD, Warnock GI, Haider A, Bengs S, Cartolano G, et al. [(11)C]mHED PET follows a two-tissue compartment model in mouse myocardium with norepinephrine transporter (NET)-dependent uptake, while [(18)F]LMI1195 uptake is NET-independent. EJNMMI Res. 2020;10:114. https://doi.org/10.1186/s13550-020-00700-7.
Froklage FE, Boellaard R, Bakker E, Hendrikse NH, Reijneveld JC, Schuit RC, et al. Quantification of 11C-Laniquidar Kinetics in the Brain. J Nucl Med. 2015;56:1730–5. https://doi.org/10.2967/jnumed.115.157586.
El Arfani C, De Veirman K, Maes K, De Bruyne E, Menu E. Metabolic Features of Multiple Myeloma. Int J Mol Sci. 2018:19. https://doi.org/10.3390/ijms19041200.
Stong N, Ortiz Estevez M, Towfic F, Samur MK, Agarwal A, Corre J, et al. Location of the t(4;14) translocation breakpoint within the NSD2 gene identifies a subset of high-risk NDMM patients. Blood. 2022; https://doi.org/10.1182/blood.2022016212.
Cavo M, Terpos E, Nanni C, Moreau P, Lentzsch S, Zweegman S, et al. Role of (18)F-FDG PET/CT in the diagnosis and management of multiple myeloma and other plasma cell disorders: a consensus statement by the International Myeloma Working Group. Lancet Oncol. 2017;18:e206–e17. https://doi.org/10.1016/S1470-2045(17)30189-4.
Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. https://doi.org/10.1016/j.cmet.2015.12.006.
Yeung J, Chang H. Genomic aberrations and immunohistochemical markers as prognostic indicators in multiple myeloma. J Clin Pathol. 2008;61:832–6. https://doi.org/10.1136/jcp.2007.049585.
Dreux N, Marty M, Chibon F, Velasco V, Hostein I, Ranchere-Vince D, et al. Value and limitation of immunohistochemical expression of HMGA2 in mesenchymal tumors: about a series of 1052 cases. Mod Pathol. 2010;23:1657–66. https://doi.org/10.1038/modpathol.2010.174.
Funding
This work was supported in part by the National Natural Science Foundation of China (Grant No.82102089 and Grant No.82171972), National Key Research and Development Program of China (Grant No. 2020YFA0909000), Hospital Development Center of Shanghai for Research called “Three-year action plan for major clinical research program” (Grant No. SHDC2020CR20708), Construction Project of Shanghai Key Laboratory of Molecular Imaging (Grant No. 18DZ2260400), and Shanghai Municipal Education Commission (Class II Plateau Disciplinary Construction Program of Medical Technology of SUMHS, 2018–2020).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Jiajin Li, Yumei Chen, and Yue Gu. The first draft of the manuscript was written by Jiajin Li and Xiaofeng Yu, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have declared that no competing interest exists.
Ethics approval
All procedures performed in studies involving human participants were approved by the Institutional Review Board of Shanghai Jiao Tong University-affiliated Ren Ji Hospital and with the principles of the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. This article does not describe any studies with animals performed by any of the authors.
Consent to participate
Written informed consent was obtained from the parents.
Consent for publication
The authors affirm that human research participants provided informed consent for publication.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Hematology.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, J., Ni, B., Yu, X. et al. Metabolic kinetic modeling of [11C]methionine based on total-body PET in multiple myeloma. Eur J Nucl Med Mol Imaging 50, 2683–2691 (2023). https://doi.org/10.1007/s00259-023-06219-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-023-06219-y