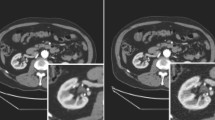
Abstract
Background
For PET/CT, the CT transmission data are used to correct the PET emission data for attenuation. However, subject motion between the consecutive scans can cause problems for the PET reconstruction. A method to match the CT to the PET would reduce resulting artifacts in the reconstructed images.
Purpose
This work presents a deep learning technique for inter-modality, elastic registration of PET/CT images for improving PET attenuation correction (AC). The feasibility of the technique is demonstrated for two applications: general whole-body (WB) imaging and cardiac myocardial perfusion imaging (MPI), with a specific focus on respiratory and gross voluntary motion.
Materials and methods
A convolutional neural network (CNN) was developed and trained for the registration task, comprising two distinct modules: a feature extractor and a displacement vector field (DVF) regressor. It took as input a non-attenuation-corrected PET/CT image pair and returned the relative DVF between them—it was trained in a supervised fashion using simulated inter-image motion. The 3D motion fields produced by the network were used to resample the CT image volumes, elastically warping them to spatially match the corresponding PET distributions. Performance of the algorithm was evaluated in different independent sets of WB clinical subject data: for recovering deliberate misregistrations imposed in motion-free PET/CT pairs and for improving reconstruction artifacts in cases with actual subject motion. The efficacy of this technique is also demonstrated for improving PET AC in cardiac MPI applications.
Results
A single registration network was found to be capable of handling a variety of PET tracers. It demonstrated state-of-the-art performance in the PET/CT registration task and was able to significantly reduce the effects of simulated motion imposed in motion-free, clinical data. Registering the CT to the PET distribution was also found to reduce various types of AC artifacts in the reconstructed PET images of subjects with actual motion. In particular, liver uniformity was improved in the subjects with significant observable respiratory motion. For MPI, the proposed approach yielded advantages for correcting artifacts in myocardial activity quantification and potentially for reducing the rate of the associated diagnostic errors.
Conclusion
This study demonstrated the feasibility of using deep learning for registering the anatomical image to improve AC in clinical PET/CT reconstruction. Most notably, this improved common respiratory artifacts occurring near the lung/liver border, misalignment artifacts due to gross voluntary motion, and quantification errors in cardiac PET imaging.
Graphical Abstract










Similar content being viewed by others
Change history
25 March 2023
A Correction to this paper has been published: https://doi.org/10.1007/s00259-023-06199-z
References
Xiao H, Ren G, Cai J. A review on 3D deformable image registration and its application in dose warping. Chin J Radiol Med Prot. 2020;1(4):171–8. https://doi.org/10.1016/j.radmp.2020.11.002.
Fu Y, et al. Deep learning in medical image registration: a review. Physics in Medicine & Biology. 2020;65(20):20TR01.
Boveiri HR, et al. Medical image registration using deep neural networks: a comprehensive review. Comput Electr Eng. 2020;87:106767.
Cao X, et al. Deep learning based inter-modality image registration supervised by intra-modality similarity. In: International workshop on machine learning in medical imaging: Springer; 2018.
Sun L, Zhang S. Deformable MRI-ultrasound registration using 3D convolutional neural network. In: Simulation, Image Processing, and Ultrasound Systems for Assisted Diagnosis and Navigation: Springer; 2018. p. 152–8.
Roy S, et al. MR to CT registration of brains using image synthesis. In: Medical Imaging 2014: Image Processing: International Society for Optics and Photonics; 2014.
Chen M, et al. Cross contrast multi-channel image registration using image synthesis for MR brain images. Med Image Anal. 2017;36:2–14.
Yu, H., et al. Learning 3D non-rigid deformation based on an unsupervised deep learning for PET/CT image registration. In Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging. 2019. International Society for Optics and Photonics.
Yu H, et al. Unsupervised 3D PET-CT image registration method using a metabolic constraint function and a multi-domain similarity measure. IEEE Access. 2020;8:63077–89.
Kang H, et al. An optimized registration method based on distribution similarity and DVF smoothness for 3D PET and CT images. IEEE Access. 2019;8:1135–45.
Jaderberg M, Simonyan K, Zisserman A. Spatial transformer networks. Adv Neural Inf Proces Syst. 2015;28:2017–25.
de Vos BD, et al. A deep learning framework for unsupervised affine and deformable image registration. Med Image Anal. 2019;52:128–43.
Balakrishnan G, et al. Voxelmorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging. 2019;38(8):1788–800.
Chee, E. and Z. Wu, Airnet: self-supervised affine registration for 3d medical images using neural networks. arXiv preprint arXiv:1810.02583, 2018.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention: Springer; 2015.
Kingma, D.P. and J. Ba, Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014.
Gould KL, et al. Frequent diagnostic errors in cardiac PET/CT due to misregistration of CT attenuation and emission PET images: a definitive analysis of causes, consequences, and corrections. J Nucl Med. 2007;48(7):1112–21.
Acknowledgements
The authors would like to thank Drs. Ian Armstrong and Rob deKemp for sharing cardiac subject data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Conflict of interest
The authors JS, VS, CH, and SV are full-time employees of Siemens Medical Solutions USA. No other potential conflicts of interest relevant to this article exist.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence).
The original online version of this article was revised due to a retrospective Open Access cancellation.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Schaefferkoetter, J., Shah, V., Hayden, C. et al. Deep learning for improving PET/CT attenuation correction by elastic registration of anatomical data. Eur J Nucl Med Mol Imaging 50, 2292–2304 (2023). https://doi.org/10.1007/s00259-023-06181-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-023-06181-9