Abstract
Purpose
Inter-subject covariance of regional 18F-fluorodeoxyglucose (FDG) PET measures (FDGcov) as proxy of brain connectivity has been gaining an increasing acceptance in the community. Yet, it is still unclear to what extent FDGcov is underlied by actual structural connectivity via white matter fiber tracts. In this study, we quantified the degree of spatial overlap between FDGcov and structural connectivity networks.
Methods
We retrospectively analyzed neuroimaging data from 303 subjects, both patients with suspected neurodegenerative disorders and healthy individuals. For each subject, structural magnetic resonance, diffusion tensor imaging, and FDG-PET data were available. The images were spatially normalized to a standard space and segmented into 62 anatomical regions using a probabilistic atlas. Sparse inverse covariance estimation was employed to estimate FDGcov. Structural connectivity was measured by streamline tractography through fiber assignment by continuous tracking.
Results
For the whole brain, 55% of detected connections were found to be convergent, i.e., present in both FDGcov and structural networks. This metric for random networks was significantly lower, i.e., 12%. Convergent were 80% of intralobe connections and only 30% of interhemispheric interlobe connections.
Conclusion
Structural connectivity via white matter fiber tracts is a relevant substrate of FDGcov, underlying around a half of connections at the whole brain level. Short-range white matter tracts appear to be a major substrate of intralobe FDGcov connections.
Similar content being viewed by others
Introduction
In the last decade, brain connectivity has evolved as a hot topic of neuroscience. Along with functional magnetic resonance imaging (fMRI), positron emission tomography (PET) with 18F-fluorodeoxyglucose (FDG) represents a valuable tool for exploring neural function in vivo. Of note, FDG-PET can also provide information on brain connectivity. The term metabolic connectivity refers to interrelations between metabolic (FDG) measurements in different brain regions [1]. This approach was shown to yield valuable knowledge on substrates of cognitive reserve [2, 3], working memory [4, 5], impulse control [6], as well as on pathophysiology and diagnosis of neurodegenerative [7,8,9,10] and non-neurodegenerative disorders [11, 12]. However, it is still unclear how MC is related to actual structural connectivity, i.e., connectivity through white matter fiber tracts. The latter can be measured in the living human brain using diffusion tensor imaging (DTI). Notably, a few DTI studies have investigated structural substrates of functional connectivity from fMRI data. Measures of structural and functional connectivity were found to be interrelated, and structurally connected cortical regions exhibited a stronger and more consistent functional connectivity than structurally unconnected regions [13]. The goal of the present study was to quantify the degree of spatial overlap between FDGcov and structural connectivity networks. We now prefer the term FDGcov, inter-subject covariance of regional FDG-PET measures, over the term metabolic connectivity to avoid a confusion with connectivity measures from dynamic/functional PET acquisitions [14]. To this end, we analyzed data from a large, heterogeneous pool of 303 subjects, both patients with suspected neurodegenerative disorders and healthy individuals.
Material and methods
Subjects
We retrospectively analyzed a database of healthy individuals and subjects who were referred to our department as part of a diagnostic work-up for a suspected neurodegenerative disorder. In total, 303 subjects whose structural MRI (sMRI), DTI, and FDG-PET data were available were included. The cohort consisted mainly of patients with mild cognitive impairment [15] and dementia [16], as well as patients with another or unspecified syndrome diagnosis, and healthy individuals. Healthy subjects were recruited mainly via advertisements in local newspapers. They had no history and symptoms of psychiatric and neurologic disorders, no complaints about cognitive impairment (n = 36) or complaints that were not confirmed on neuropsychological testing (n = 4). Demographic data of the cohort according to a syndrome diagnosis are summarized in Table 1.
The study was carried out in accordance with the latest version of the Declaration of Helsinki, after the consent procedures had been approved by the local ethics committee. Written informed consent was obtained from all subjects or their legal representatives.
Image data acquisition
Imaging data were acquired on a fully integrated Siemens Biograph mMR (Siemens Medical Solutions, Knoxville, USA) PET/MR system [17]. PET data were acquired in list mode over 15 min, 30 min after an intravenous injection of approximately 185 MBq 18F-FDG. A Dixon T1 MRI sequence was run in parallel with PET to ensure optimal temporal and regional correspondence between two modalities for later attenuation correction. DTI data were acquired using a fast gradient echo-planar imaging sequence with a TE of 82 ms, a TR of 12,100 ms, and a flip angle of 90°. Per subject, 30 volumes with b = 800 s/mm2 and distinct diffusion-encoding directions and one volume with b = 0 s/mm2 were acquired. Images had a field of view of 208 mm with 130 × 130 image matrix and 2 mm slice thickness. A high-resolution structural MRI sequence (T1-weighted MPRAGE) was acquired for anatomical correspondence. PET emission data were corrected for random coincidences, dead time, scatter, and attenuation. Resulting sinograms were reconstructed using a filtered back-projection algorithm (FORE + FBP, Siemens syngo MR B18P) with a 5-mm Hamming filter into 192 × 192 × 128 volumes at a field of view of 450 mm. The voxel size was 3.7 × 3.7 × 2.3 mm3.
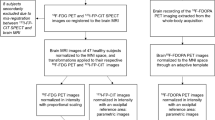
Preprocessing of image data
The accuracy of the alignment between MRI and PET images was visually inspected in PMOD (PMOD Technologies LLC, CH). The MPRAGE images were then spatially normalized to the Montreal Neurological Institute space [18] using a simultaneous tissue segmentation/spatial normalization tool in SPM12 (Wellcome Trust Centre for Neuroimaging, UCL, London). Resulting gray matter maps were thresholded with a probability value of 0.5. Transformation matrices were then applied to corresponding DTI and PET images. No smoothing was applied. The spatially normalized PET images were then parcellated into 62 non-overlapping regions (region volume ≥ 1 cm3) according to the Hammers atlas [19]. The PET images were corrected for partial-volume effects using a voxel-wise approach [20] implemented in PETPVC (Dept. of Nuclear Medicine, UCL). The regional values were scaled using proportional scaling to the mean value of the whole gray matter (segmented from T1 MRI). Although this approach is not optimal for univariate analyses [21,22,23], it may provide more stable results in multivariate analyses as those utilized here ([24], own unpublished data). DTI volumes were corrected for eddy currents and head motion using FSL [25]. The general processing of the data was performed using Python programming language with the nipype library [26, 27].
FDG cov network
A sparse inverse covariance estimation (SICE) method established in our previous studies [5, 8] was employed to calculate FDGcov. The FDG PET data are represented as \(\mathbf{\rm X}\in {\mathbb{R}}^{n\times m}\), where m denotes the number of anatomical regions and n is the number of scanned subjects. Thus, \({\mathbf{x}}_{i}, 1\le i\le n\) is a m-dimensional vector for subject i and is assumed to follow a multivariate Gaussian distribution \({\mathbf{x}}_{i} \sim \mathcal{N}({\varvec{\upmu}},{\varvec{\Sigma}})\) [8], where \({\varvec{\upmu}}\in {\mathbb{R}}^{m}\) is a mean vector, and \({\varvec{\Sigma}}\in {\mathbb{R}}^{m\times m}\) is an underlying covariance matrix. A sample covariance matrix of the FDG data is \(\widehat{{\varvec{\Sigma}}}\), and the inverse covariance matrix is estimated as following:
where λ is a regularization parameter that regularizes the sparsity, and \({\Vert \Vert }_{1}\) is the L1 norm. This is a LASSO model [28] which keeps \({\varvec{\Theta}}\) sparse: the regularization parameter λ directly controls how many connections will be identified, i.e., how many entries will be nonzero. This estimated sparse inverse covariance matrix \({\varvec{\Theta}}\) is treated as FDGcov network thereafter. Only positive entries in the sparse connectivity pattern were considered as connections, resulting in binary matrices [29].
Structural connectivity network
Camino [30] was employed to fit the diffusion tensors with logistic regression [31] and to perform streamline tractography through fiber assignment by continuous tracking (FACT, [32]). The algorithm was seeded with all white-matter voxels and stopped when reaching a voxel with a fractional anisotropy (FA) value of less than 0.2 or when the curvature of a tract exceeded 50° over 5 mm [33, 34]. From the tractography results, the mean fractional anisotropy (FA) of a given fiber tract (between the ROIs above, if detected) across subjects was implemented in structural connectivity matrices.
Network efficiency
Network efficiency (NE) analyses were conducted to define a reasonable number of connections for quantification of the network overlap. NE characterizes information transfer in a network [35]. In particular, local efficiency (LE) describes a network’s resistance to failure on a small scale, e.g., when a node is removed. As compared to global efficiency, LE is not proportional to the number of connections [36], making it more suitable for the above purpose. The network was modeled as a simple graph of m vertices (node) and \({n}_{connected}\) edges (connections). Herewith, a node in the network corresponds to a specific anatomical region. For two network nodes \(k\) and \(l\), the shortest path length \({d}_{kl}\) is the number of edges on the shortest path. For each node i, a subnetwork Gi is defined as a neighborhood subgraph of this node. The efficiency of the subnetwork Gi is defined as average inverse of the shortest path lengths \({d}_{kl}\) in this subnetwork. The local efficiency \({E}_{local}\) then averages the efficiencies across the subnetworks of all nodes:
LE is a scaled measure ranging from 0 to 1, with a value of 1 indicating maximum LE in the network. Conceptually, a high LE represents an effective information transfer within their immediate local communities, enabling effective information processing in the network. The LE of a comparable random network was employed as reference. Specifically, random connectivity matrices with the same number of connections and degree of distribution were generated using a random rewiring method [35, 37]. Given that the LE from a random matrix increases with the number of connections [38], the genuine LE (gLE) was employed to eliminate the influence of random effects. It was defined by subtracting the expected LE of the randomly rewired networks (mean of bootstrapping results) from the original LE of the corresponding FDGcov and structural networks. Following the theory of signal processing [39], we selected a cutoff value at half maximum of gLE to define a focus window for the number of connections (full-width at half maximum) for further analyses. To assess the stability of results across different sampling populations, 100 bootstrap samples were generated by random resampling with replacement.
Overlap between FDG cov and structural networks
Similar to previous studies, sparsity-based thresholding was employed to restrict the number of connections in both networks [35]. Networks with the same number of connections \({n}_{connected}\) according to the NE analysis were generated for pattern comparison. A threshold at tract-averaged FA values was determined such that only the \({n}_{connected}\) strongest structural connections were left. For FDGcov, a scalar regularization parameter λ of SICE was chosen such that \({n}_{connected}\) entries were nonzero above the diagonal of the resulting inverse covariance matrix [29]. Diagonal elements, representative of self-connections, were ignored to increase the robustness of the regularization.
A convergence ratio (CR) is defined by dividing the number of the pairs which are connected in both networks (\({n}_{convergent}\)) by the number of present connections (\(n_{connected}\)),
This is equivalent to sensitivity in binary classification if the presence of a connection is treated as positive class and is also equivalent to so called Dice similarity coefficient. For individual hemispheres, CR was calculated at an unequal number of connections in two networks, and it is adapted to the following expression:
where \({n}_{metabolic}\) is the number of functional connections and \({n}_{structural}\) is the number of structural connections. CR was analyzed as a function of the number of connections \({n}_{connected}.\) As reference, randomly rewired matrices of the FDGcov and structural networks with the same number of connections were generated 100 times. Figure 1 summarizes the pipeline of the PET and DTI image analyses.
Results
Figure 2 shows LE of FDGcov and structural networks at different numbers of connections. Both networks appeared to have a significantly higher LE compared to the randomly generated networks (FDGcov network: p = 1.3 × 10−178 and structural network: p = 9.4 × 10−201). The FDGcov network had a higher variation in the bootstrapped networks (p = 2.4 × 10−174) as well as lower LE (p = 2.6 × 10−158) than the structural network. In the range of 65–338 connections, both FDGcov and structural networks had a gLE above the half of maximum, such that this range was used for visualization purposes (Fig. 3). A plateau (> 90% of the maximum) of the cumulative gLE of FDGcov and structural connectivity corresponded to the range of connections 128–275 (Fig. 2C). Thus, we used this range in further quantitative analyses.
Network efficiency: Local efficiency for FDGcov (A) and structural (B) connectivity; real data—red line, results of bootstrapping—blue line, and results of randomly generated networks—black line. C Genuine local efficiency for FDGcov—green line, structural connectivity—red line, and cumulative (summed)—black line. The range of half maximum of FDGcov and structural connectivity networks is illustrated as dashed lines in green and red, respectively. The black dashed line illustrates the 0.9 of maximum total genuine local efficiency, reflecting the optimal number of connections for both networks
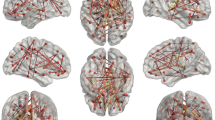
CR of the networks was significantly higher (p = 4.9 × 10−321) than that by chance (Fig. 3). In the plateau window, i.e., at 128–275 connections, the average CR was 0.54 ± 0.03 (0.47–0.57), while CR of the random networks was 0.11 ± 0.03 (0.01–0.21).. The results of bootstrapping did not perfectly overlap with the real data results. That is, in Fig. 3, the red line does not exactly follow the solid blue line, likely due to numerical instability of SICE in the process of resampling. Figure 4 depicts matrices of FDGcov and structural networks at \({n}_{connected}=215\), i.e., the maximum of cumulative gLE (Fig. 2C). These networks are depicted in Fig. 5. The proportion of interhemispheric connections was 23% (n = 49) and 15% (n = 32) in FDGcov and structural networks, respectively. Among intrahemispheric connections, the proportion of intralobe connections was 50% (n = 83) and 45.9% (n = 84), respectively. Quantitative results for the spatial overlap are summarized in Table 2. CR for intralobe and interlobe connections was 0.80 and 0.37, respectively (p = 6 × 10−16, Wilcoxon rank test). There was no remarkable difference between the hemispheres (Supplemental Table 2). CR values for random networks are summarized in Supplemental Table 3. For the whole brain CR was 0.12.
Matrix visualizations of connectivity between 62 regions with 215 connections. FDGcov (green), structural (red), and convergent (black) connections. Boxes capture anatomically related regions, i.e., within (from top to the bottom) the frontal, temporal, parietal, and occipital lobes, subcortical and limbic regions. For regional labels see Supplemental Table 1
Discussion
The present study examined the degree of spatial overlap between FDGcov and structural connectivity. The overlap appeared to be significantly higher for the FDGcov and structural networks than for random ones. At the whole brain level CR was 55%. In other words, around a half of detected FDGcov connections in the brain appear to be underlied by white matter tracts. Furthermore, 80% of intralobe FDGcov connections that are supposed to reflect short-range connections [40] were found to have a structural substrate. These results support FDGcov as a sovereign index of brain connectivity.
While a few groups have studied the relationship between fMRI functional and DTI structural connectivity [13, 41, 42], there are still no quantitative data on their spatial overlap. Nevertheless, in line with our data, strong intrahemispheric [43] and weaker transmodal (e.g., interhemispheric) structure–function correlations [44] have been reported. Moreover, a so-called spatial proximity was suggested to be a major determinant of structure–function relationships in diffusion imaging and fMRI data [45]. We found that the proportion of different types of FDGcov connections followed the order of structural ones, i.e., intrahemispheric-intralobe > intrahemispheric-interlobe > interhemispheric-interlobe. These results are in line with DTI studies. In fact, short-range connections were shown to by far outnumber long-range connections passing through corpus callosum [46, 47]. Further, structural connections within an anatomical region were found to be more common than those between regions, followed by interhemispheric connections [48]. However, we detected somewhat more interhemispheric connections in the PET than in the DTI data, i.e., 23% vs. 15%. This is likely related to differences in connectivity modeling. While tractography captures connections underlying an anatomical network of axonal fibers [49], FDGcov modeling “detects” indirect connections if two regions have a similar level of FDG uptake [29, 50]. Given a relative symmetry in cerebral glucose metabolism [51, 52], there is a substantial likelihood that homotopic regions of two hemispheres appear to be connected in FDG-PET data, even though being unconnected anatomically. A similar observation was made by numerous fMRI studies, where strong functional connectivity was found between homotopic regions that are known to be unconnected anatomically (for a review, see Suárez et al. [53]).
While indirect connections in FDG-PET data may be considered as false positive, false negative connections are likely in DTI data. Specifically, deterministic tractography, as used in the present study, terminates streamlines in voxels with sub-threshold FA values. Hence, it was suggested that this tracking approach might miss particularly weak long-range connections (i.e., low FA value at a voxel level) [46, 54]. In the same vein, Sinke et al. reported a relatively high rate of false negative tractography reconstructions for long-range connected cortical areas, as validated against neuronal tracer connectivity measures in the rat [55]. Similarly, post-mortem invasive tracer studies in macaques found that false negatives exhibited a significantly larger connection distance than false positives or true positives [56]. In the present study, the Euclidean distance between any couple of regions of interest from the Hammers atlas negatively correlated with the number of streamlines between the same regions (data not shown), supporting the above notion. Although probabilistic tractography detects so-called kissing fibers in a more sensitive fashion, it seems not advantageous regarding long-range connections [57]. Future tractography studies should test a range of FA thresholds [55].
As a major study limitation, we utilized a heterogeneous cohort that included both neurodegenerative and non-neurodegenerative entities. An unverified assumption behind this pragmatic approach is that a disease equivalently affects FDGcov and structural connectivity. Still, the overlap might differ in the healthy state and in a disease. Further studies should address the impact of data heterogeneity and sample size on estimates of FDGcov in general and on the overlap between FDGcov and structural connectivity in particular. Moreover, a differentiated analysis at the lobe level may produce novel insights into the structural substrates of FDGcov.
Conclusion
Structural connectivity via white matter tracts is a relevant substrate of FDGcov, underlying around a half of connections at the whole brain level. Short-range white matter tracts are a major substrate of intralobe FDGcov connections. The present study represents the first valuable reference on structural substrates of FDGcov, contributing to establishment of FDGcov as a sovereign index of brain connectome.
Data availability
Data will be made available upon request.
Code availability
Code will be made available upon request.
References
Yakushev I, Drzezga A, Habeck C. Metabolic connectivity: methods and applications. Curr Opin Neurol. 2017;30:677–85.
Morbelli S, Perneczky R, Drzezga A, Frisoni GB, Caroli A, van Berckel BNM, et al. Metabolic networks underlying cognitive reserve in prodromal Alzheimer disease: a European Alzheimer disease consortium project. J Nucl Med. 2013;54:894–902.
Perani D, Farsad M, Ballarini T, Lubian F, Malpetti M, Fracchetti A, et al 2017 The impact of bilingualism on brain reserve and metabolic connectivity in Alzheimer’s dementia. PNAS [Internet]. National Academy of Sciences [cited 2020 Sep 22]; Available from: https://www.pnas.org/content/early/2017/01/24/1610909114
Yakushev I, Chételat G, Fischer FU, Landeau B, Bastin C, Scheurich A, et al 2013 Metabolic and structural connectivity within the default mode network relates to working memory performance in young healthy adults. NeuroImage [Internet]. [cited 2020 Mar 25];79:184–90. Available from: http://www.sciencedirect.com/science/article/pii/S1053811913004163
Zou N, Chetelat G, Baydogan MG, Li J, Fischer FU, Titov D, et al. Metabolic connectivity as index of verbal working memory. J Cereb Blood Flow Metab. 2015;35:1122–6.
Verger A, Klesse E, Chawki MB, Witjas T, Azulay J-P, Eusebio A, et al. Brain PET substrate of impulse control disorders in Parkinson’s disease: a metabolic connectivity study. Hum Brain Mapp. 2018;39:3178–86.
Sala A, Caminiti SP, Presotto L, Premi E, Pilotto A, Turrone R, et al 2017 Altered brain metabolic connectivity at multiscale level in early Parkinson’s disease. Scientific Reports [Internet]. Nature Publishing Group [cited 2020 Mar 25];7:1–12. Available from: https://www.nature.com/articles/s41598-017-04102-z
Titov D, Diehl-Schmid J, Shi K, Perneczky R, Zou N, Grimmer T, et al. Metabolic connectivity for differential diagnosis of dementing disorders. J Cereb Blood Flow Metab. 2017;37:252–62.
Caminiti SP, Tettamanti M, Sala A, Presotto L, Iannaccone S, Cappa SF, et al. Metabolic connectomics targeting brain pathology in dementia with Lewy bodies. J Cereb Blood Flow Metab. 2017;37:1311–25.
Huber M, Beyer L, Prix C, Schönecker S, Palleis C, Rauchmann B-S, et al. Metabolic correlates of dopaminergic loss in dementia with Lewy bodies. Mov Disord. 2020;35:595–605.
Kim H, Kim YK, Lee JY, Choi AR, Kim DJ, Choi J-S 2019 Hypometabolism and altered metabolic connectivity in patients with internet gaming disorder and alcohol use disorder. Progress in Neuro-Psychopharmacology and Biological Psychiatry [Internet]. [cited 2020 Sep 22];95:109680. Available from: http://www.sciencedirect.com/science/article/pii/S0278584619301952
Shim H-K, Lee H-J, Kim SE, Lee BI, Park S, Park KM 2020 Alterations in the metabolic networks of temporal lobe epilepsy patients: a graph theoretical analysis using FDG-PET. NeuroImage: Clinical [Internet]. [cited 2020 Sep 22];27:102349. Available from: http://www.sciencedirect.com/science/article/pii/S2213158220301868
Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, et al 2009 Predicting human resting-state functional connectivity from structural connectivity. PNAS [Internet]. National Academy of Sciences [cited 2020 Aug 13];106:2035–40. Available from: https://www.pnas.org/content/106/6/2035
Jamadar SD, Ward PGD, Liang EX, Orchard ER, Chen Z, Egan GF. Metabolic and hemodynamic resting-state connectivity of the human brain: a high-temporal resolution simultaneous BOLD-fMRI and FDG-fPET multimodality study. Cereb Cortex. 2021;31:2855–67.
Winblad B, Palmer K, Kivipelto M, Jelic V, Fratiglioni L, Wahlund L-O, et al 2004 Mild cognitive impairment – beyond controversies, towards a consensus: report of the International Working Group on Mild Cognitive Impairment. Journal of Internal Medicine [Internet]. [cited 2021 Sep 23];256:240–6. Available from: https://onlinelibrary.wiley.com/doi/abs/https://doi.org/10.1111/j.1365-2796.2004.01380.x
World Health Organization 1993 The ICD-10 classification of mental and behavioural disorders: diagnostic criteria for research. CIM-10/ICD-10: Classification internationale des maladies Dixième révision Chapitre V(F): troubles mentaux et troubles du comportement: critères diagnostiques pour la recherche [Internet]. Geneva: World Health Organization Available from: https://apps.who.int/iris/handle/10665/37108
Delso G, Fürst S, Jakoby B, Ladebeck R, Ganter C, Nekolla SG, et al 2011 Performance measurements of the Siemens mMR integrated whole-body PET/MR scanner. J Nucl Med [Internet]. Society of Nuclear Medicine [cited 2020 Aug 13];52:1914–22. Available from: http://jnm.snmjournals.org/content/52/12/1914
Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, et al 2001 A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci [Internet]. [cited 2020 Aug 13];356:1293–322. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1088516/
Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19:224–47.
Erlandsson K, Buvat I, Pretorius PH, Thomas BA, Hutton BF 2012 A review of partial volume correction techniques for emission tomography and their applications in neurology, cardiology and oncology. Phys Med Biol [Internet]. IOP Publishing [cited 2020 Aug 13];57:R119–59. Available from: https://doi.org/10.1088/2F0031-9155/2F57/2F21/2Fr119
Borghammer P, Aanerud J, Gjedde A. Data-driven intensity normalization of PET group comparison studies is superior to global mean normalization. Neuroimage. 2009;46:981–8.
Yakushev I, Landvogt C, Buchholz H-G, Fellgiebel A, Hammers A, Scheurich A, et al. Choice of reference area in studies of Alzheimer’s disease using positron emission tomography with fluorodeoxyglucose-F18. Psychiatry Res. 2008;164:143–53.
Yakushev I, Hammers A, Fellgiebel A, Schmidtmann I, Scheurich A, Buchholz H-G, et al. SPM-based count normalization provides excellent discrimination of mild Alzheimer’s disease and amnestic mild cognitive impairment from healthy aging. Neuroimage. 2009;44:43–50.
Spetsieris PG, Eidelberg D. Scaled subprofile modeling of resting state imaging data in Parkinson’s disease: methodological issues. Neuroimage. 2011;54:2899–914.
Andersson JLR, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–78.
Gorgolewski K, Burns CD, Madison C, Clark D, Halchenko YO, Waskom ML, et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front Neuroinform. 2011;5:13.
Savio AM, Schutte M, Graña M, Yakushev I 2017 Pypes: Workflows for processing multimodal neuroimaging data. Front Neuroinform [Internet]. [cited 2018 Jun 8];11. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5387693/
Tibshirani R 1996 Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society: Series B (Methodological) [Internet]. [cited 2020 Oct 22];58:267–88. Available from: https://rss.onlinelibrary.wiley.com/doi/abs/https://doi.org/10.1111/j.2517-6161.1996.tb02080.x
Huang S, Li J, Sun L, Ye J, Fleisher A, Wu T, et al. Learning brain connectivity of Alzheimer’s disease by sparse inverse covariance estimation. Neuroimage. 2010;50:935–49.
Cook PA, Bai Y, Nedjati-Gilani S, Seunarine KK, Hall MG, Parker GJ, et al 2006 Camino: open-source diffusion-MRI reconstruction and processing. in 14th scientific meeting of the International Society for Magnetic Resonance in Medicine Seattle, WA, USA p 2759. 2006;1.
Basser PJ, Mattiello J, Lebihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. Journal of Magnetic Resonance, Series B [Internet]. 1994 [cited 2020 Aug 13];103:247–54. Available from: http://www.sciencedirect.com/science/article/pii/S1064186684710375
Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 1999;45:265–9.
Thiebaut de Schotten M, Ffytche DH, Bizzi A, Dell’Acqua F, Allin M, Walshe M, et al. Atlasing location, asymmetry and inter-subject variability of white matter tracts in the human brain with MR diffusion tractography. Neuroimage. 2011 54:49–59.
Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, et al. Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage [Internet]. 2007 [cited 2020 Sep 23];36:630–44. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2350213/
Gong G, He Y, Chen ZJ, Evans AC. Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. Neuroimage. 2012;59:1239–48.
Koenis MMG, Brouwer RM, van den Heuvel MP, Mandl RCW, van Soelen ILC, Kahn RS, et al. Development of the brain’s structural network efficiency in early adolescence: a longitudinal DTI twin study. Hum Brain Mapp. 2015;36:4938–53.
Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–3.
Achard S, Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Comput Biol [Internet]. 2007 [cited 2017 Nov 28];3. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1794324/
Freyer F, Aquino K, Robinson PA, Ritter P, Breakspear M. Bistability and non-Gaussian fluctuations in spontaneous cortical activity. J Neurosci [Internet]. Society for Neuroscience; 2009 [cited 2020 Aug 13];29:8512–24. Available from: https://www.jneurosci.org/content/29/26/8512
Ouyang M, Kang H, Detre JA, Roberts TPL, Huang H. Short-range connections in the developmental connectome during typical and atypical brain maturation. Neurosci Biobehav Rev [Internet]. 2017 [cited 2020 Aug 14];83:109–22. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5730465/
Straathof M, Sinke MR, Dijkhuizen RM, Otte WM. A systematic review on the quantitative relationship between structural and functional network connectivity strength in mammalian brains. J Cereb Blood Flow Metab. 2019;39:189–209.
Zimmermann J, Ritter P, Shen K, Rothmeier S, Schirner M, McIntosh AR. Structural architecture supports functional organization in the human aging brain at a regionwise and network level. Hum Brain Mapp. 2016;37:2645–61.
Uddin LQ, Supekar KS, Ryali S, Menon V. Dynamic reconfiguration of structural and functional connectivity across core neurocognitive brain networks with development. J Neurosci [Internet]. Society for Neuroscience; 2011 [cited 2020 Aug 14];31:18578–89. Available from: https://www.jneurosci.org/content/31/50/18578
Roland JL, Snyder AZ, Hacker CD, Mitra A, Shimony JS, Limbrick DD, et al. On the role of the corpus callosum in interhemispheric functional connectivity in humans. PNAS [Internet]. National Academy of Sciences; 2017 [cited 2020 Aug 14];114:13278–83. Available from: https://www.pnas.org/content/114/50/13278
Vázquez-Rodríguez B, Suárez LE, Markello RD, Shafiei G, Paquola C, Hagmann P, et al. Gradients of structure–function tethering across neocortex. PNAS [Internet]. National Academy of Sciences; 2019 [cited 2020 Aug 14];116:21219–27. Available from: https://www.pnas.org/content/116/42/21219
Markov NT, Misery P, Falchier A, Lamy C, Vezoli J, Quilodran R, et al. Weight consistency specifies regularities of macaque cortical networks. Cereb Cortex. 2011;21:1254–72.
Schüz A, Braitenberg V, Schüz R. Miller A. The human cortical white matter: quantitative aspects of cortico-cortical long-range connectivity. Cortical areas: unity and diversity [Internet]. 2002 [cited 2019 Aug 29];377–85. Available from: https://pure.mpg.de/pubman/faces/ViewItemFullPage.jsp?itemId=item_1792865_1
Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, et al. Mapping the structural core of human cerebral cortex. PLOS Biology [Internet]. 2008 [cited 2019 Aug 30];6:e159. Available from: https://journals.plos.org/plosbiology/article?id=https://doi.org/10.1371/journal.pbio.0060159
Hahn G, Skeide MA, Mantini D, Ganzetti M, Destexhe A, Friederici AD, et al. A new computational approach to estimate whole-brain effective connectivity from functional and structural MRI, applied to language development. Scientific Reports [Internet]. Nature Publishing Group; 2019 [cited 2020 Sep 22];9:8479. Available from: https://www.nature.com/articles/s41598-019-44909-6
Di X, Biswal BB, Alzheimer’s Disease Neuroimaging Initiative. Metabolic brain covariant networks as revealed by FDG-PET with reference to resting-state fMRI networks. Brain Connect. 2012;2:275–83.
Amend M, Ionescu TM, Di X, Pichler BJ, Biswal BB, Wehrl HF. Functional resting-state brain connectivity is accompanied by dynamic correlations of application-dependent [18F]FDG PET-tracer fluctuations. NeuroImage [Internet]. 2019 [cited 2020 Sep 22];196:161–72. Available from: http://www.sciencedirect.com/science/article/pii/S1053811919303209
Horwitz B, Duara R, Rapoport SI. Intercorrelations of glucose metabolic rates between brain regions: application to healthy males in a state of reduced sensory input. J Cereb Blood Flow Metab. 1984;4:484–99.
Suárez LE, Markello RD, Betzel RF, Misic B. Linking structure and function in macroscale brain networks. Trends in Cognitive Sciences [Internet]. 2020 [cited 2020 Aug 14];24:302–15. Available from: http://www.sciencedirect.com/science/article/pii/S1364661320300267
Gallos LK, Makse HA, Sigman M. A small world of weak ties provides optimal global integration of self-similar modules in functional brain networks. PNAS [Internet]. National Academy of Sciences; 2012 [cited 2020 May 19];109:2825–30. Available from: https://www.pnas.org/content/109/8/2825
Sinke MRT, Otte WM, Christiaens D, Schmitt O, Leemans A, van der Toorn A, et al. Diffusion MRI-based cortical connectome reconstruction: dependency on tractography procedures and neuroanatomical characteristics. Brain Struct Funct [Internet]. 2018 [cited 2020 Sep 22];223:2269–85. Available from:https://doi.org/10.1007/s00429-018-1628-y
Li L, Rilling JK, Preuss TM, Glasser MF, Damen FW, Hu X. Quantitative assessment of a framework for creating anatomical brain networks via global tractography. Neuroimage [Internet]. 2012 [cited 2021 Mar 18];61:1017–30. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3407566/
Khalsa S, Mayhew SD, Chechlacz M, Bagary M, Bagshaw AP. The structural and functional connectivity of the posterior cingulate cortex: comparison between deterministic and probabilistic tractography for the investigation of structure-function relationships. Neuroimage. 2014;102(Pt 1):118–27.
Funding
Open Access funding enabled and organized by Projekt DEAL. Isabelle Ripp is supported by a DFG grant to Igor Yakushev (YA 373/3–1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with animals performed by any of the authors. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Conflict of interest
The authors have no competing interests relevant to this study.
Janine Diehl-Schmid has received consultant or lecture fees from Novartis and Roche Medicine, as well as grants from the German Alzheimer Society, Bavarian State Ministry of Health and Care, German Research Foundation (DFG), and Federal Ministry of Education and Research Germany (BMBF). Timo Grimmer has received consulting fees from Biogen, Bracket, Eli Lilly, Functional Neuromodulation, Iqvia/Quintiles, Novartis, Novo Nordisk, and Roche Pharma; lecture fees from Actelion, B.Braun, Biogen, Eli Lilly, Life Molecular Imaging, Novartis, Parexel, and Roche Pharma; and grants to his institution from Actelion and Novartis. Igor Yakushev has received consultant or lecture fees from Blue Earth Diagnostics, ABC-CRO, Piramal, Pentixapharm, as well as grants from the Alzheimer Research Initiative Germany, German Research Foundation (DFG), and Federal Ministry of Education and Research Germany (BMBF). Kuangyu Shi received sponsorships and grants from Novartis, PMOD, Hermes, Siemens Healthineer, PET AG Bern, and Swiss National Science Foundation (SNF).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Neurology.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yakushev, I., Ripp, I., Wang, M. et al. Mapping covariance in brain FDG uptake to structural connectivity. Eur J Nucl Med Mol Imaging 49, 1288–1297 (2022). https://doi.org/10.1007/s00259-021-05590-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-021-05590-y