Abstract
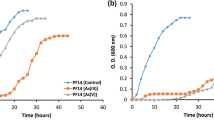
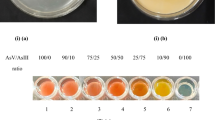
Innovative methods to lower arsenic (As) exposure are sought. The As regulatory protein (ArsR) is reported of having high affinity and specificity to arsenite [As(III)]. Rhodopseudomonas palustris CGA009 is a good model organism for studying As detoxification due to at least three ars operons and four diverse arsRRP1–4 on the genome. In this study, four Escherichia coli harboring arsRRP1–4 derived from CGA009 were engineered and tested regarding their As resistance. The results showed that E. coli (arsRRP2) displayed robust As(III) resistance, and its growth inhibition rate was only 2.9% when exposed to 3.0 mmol/L As(III). At pH 7.0, E. coli (arsRRP2) showed an enhanced As adsorption capacity. As(III) (2.32 mg/g (dry weight, dw)) and 1.47 mg/g arsenate [As(V)] was adsorbed representing a 4.2-fold and 1.3-fold increase respectively compared to the control strain. The adsorption process was well fitted to Langmuir isothermal mode. E. coli (arsRRP2) (1.0~12.0 g/L) could remove 30.3~82.2% of As (III) when exposed to 10 μg/L As(III). No increase in absorption to copper(II), zinc(II), chromium(III), and lead(II) could be detected. Our studies revealed that arsRRP1–4 from CGA009 could confer As(III) resistance; E. coli (arsRRP2) displayed the highest As resistance, selectivity, and adsorption capacity within a wider pH (5.0~9.0) and salinity (0~15.0 g/L NaCl) range, especially important as it could remove As(III) from low concentration As-containing water.





Similar content being viewed by others
References
Bereza-Malcolm L, Aracic S, Mann G, Franks AE (2018) The development and analyses of several gram-negative arsenic biosensors using a synthetic biology approach. Sensors Actuators B Chem 256:117–125
Chatterjee S, Mitra A, Datta S, Veer V (2013) Phytoremediation protocols: an overview. In: Gupta DK (ed) Plant-based remediation processes. Berlin, pp 1–18
Guo SW, Chang-Jiang L, Zhang Y, Zhao CG (2014) Diversity and evolutionary analysis of arsenic metabolism gene in R. palustris. J Huaqiao Univ 35(2):175–179
Joshi DN, Flora SJ, Kalia K (2009) Bacillus sp. strain DJ-1, potent arsenic hypertolerant bacterium isolated from the industrial effluent of India. J Hazard Mater 166(2–3):1500–1505
Kang YS, Brame K, Jetter J, Bothner BB, Wang G, Thiyagarajan S, Mcdermott TR (2016) Regulatory activities of four ArsR proteins in Agrobacterium tumefaciens 5A. Appl Environ Microbiol 82(12):3471–3480
Kostal J, Yang R, Wu CH, Mulchandani A, Chen W (2004) Enhanced arsenic accumulation in engineered bacterial cells expressing ArsR. Appl Environ Microbiol 70(8):4582–4587
Kruger MC, Bertin PN, Heipieper HJ, Arsèneploetze F (2013) Bacterial metabolism of environmental arsenic--mechanisms and biotechnological applications. Appl Microbiol Biotechnol 97(9):3827–3841
Liu Z, Rensing C, Rosen BP (2014) Resistance pathways for metalloids and toxic metals. In: Culotta V, Scott RA (eds) Metals in cells. Wiley & Sons, Hoboken, pp 429–442
Lü CJ, Zhang Y, Zhao C, Guo S, Yang S (2012) Arsenic resistance mechanisms in Rhodopseudomonas palustris under anaerobic and light conditions. Acta Sci Circumst 32(10):2375–2383
Ma Y, Lin J, Zhang C, Ren Y, Lin J (2011) Cd(II) and As(III) bioaccumulation by recombinant Escherichia coli expressing oligomeric human metallothioneins. J Hazard Mater 185(2–3):1605–1608
Mukhopadhyay R, Rosen BP, Phung LT, Silver S (2002) Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev 26(3):311–325
Ordóñez E, Thiyagarajan S, Cook JD, Stemmler TL, Gil JA, Mateos LM, Rosen BP (2008) Evolution of metal(loid) binding sites in transcriptional regulators. J Biol Chem 283(37):25706–25714
Podder MS, Majumder CB (2016) Bioaccumulation of As(III)/As(V) ions by living cells of Corynebacterium glutamicum MTCC 2745. Sep Sci Technol 51(18):2970–2990
Podder MS, Majumder CB (2017) Bioremediation of As(III) and As(V) from wastewater using living cells of Bacillus arsenicus MTCC 4380. Environ Nanotechnol Monitor Manag 8:25–47
Prasad KS, Ramanathan AL, Paul J, Subramanian V, Prasad R (2013) Biosorption of arsenite (As(+3)) and arsenate (As(+5)) from aqueous solution by Arthrobacter sp. biomass. Environ Technol 34(19):2701–2708
Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C (2006) Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci U S A 103(7):2075–2080
Qin J, Fu HL, Ye J, Bencze KZ, Stemmler TL, Rawlings DE, Rosen BP (2009) Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. J Biol Chem 282(47):34346–34355
Saha RP, Samanta S, Patra S, Sarkar D, Saha A, Singh MK (2017) Metal homeostasis in bacteria: the role of ArsR-SmtB family of transcriptional repressors in combating varying metal concentrations in the environment. Biometals 30:459–503
Sauge-Merle S, Cuiné S, Carrier P, Lecomte-Pradines C, Luu DT, Peltier G (2003) Enhanced toxic metal accumulation in engineered bacterial cells expressing Arabidopsis thaliana phytochelatin synthase. Appl Environ Microbiol 69(1):490–494
Sauge-Merle S, Lecomte-Pradines C, Carrier P, Cuiné S, Dubow M (2012) Heavy metal accumulation by recombinant mammalian metallothionein within Escherichia coli protects against elevated metal exposure. Chemosphere 88(8):918–924
Shi W, Dong J, Scott RA, Ksenzenko MY, Rosen BP (1996) The role of arsenic-thiol interactions in metalloregulation of the ars operon. J Biol Chem 271(16):9291–9297
Singh S, Mulchandani A, Chen W (2008) Highly selective and rapid arsenic removal by metabolically engineered Escherichia coli cells expressing Fucus vesiculosus metallothionein. Appl Environ Microbiol 74(9):2924–2927
Singh S, Kang SH, Lee W, Mulchandani A, Chen W (2010) Systematic engineering of phytochelatin synthesis and arsenic transport for enhanced arsenic accumulation in E. coli. Biotechnol Bioeng 105(4):780–785
Takeuchi M, Kawahata H, Gupta LP, Kita N, Morishita Y, Ono Y, Komai T (2007) Arsenic resistance and removal by marine and non-marine bacteria. J Biotechnol 127(3):434–442
Wang S, Zhao X (2009) On the potential of biological treatment for arsenic contaminated soils and groundwater. J Environ Manag 90(8):2367–2376
Xu C, Shi W, Rosen BP (1996) The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J Biol Chem 271(5):2427–2432
Xu S, Xu R, Nan Z, Chen P (2018) Bioadsorption of arsenic from aqueous solution by the extremophilic bacterium Acidithiobacillus ferrooxidans DLC-5. Biocatal Biotransfor, 1–9. doi:https://doi.org/10.1080/10242422.2018.1447566
Yang T, Liu JW, Gu C, Chen ML, Wang JH (2013) Expression of arsenic regulatory protein in Escherichia coli for selective accumulation of methylated arsenic species. ACS Appl Mater Interfaces 5(7):2767–2772
Zhao C, Zhang Y, Chan Z, Chen S, Yang S (2015) Insights into arsenic multi-operons expression and resistance mechanisms in Rhodopseudomonas palustris CGA009. Front Microbiol 6:986
Zhu YG, Yoshinaga M, Zhao FJ, Rosen BP (2014) Earth abides arsenic biotransformations. Annu Rev Earth Planet Sci 42:443–467
Funding
This study was funded by the National Marine Public Industry Research (No. 201505026), by the Natural Science Foundation of Fujian Province (No. 2018J01049, 2015 J01137), and by the Subsidized Project for Cultivating Postgraduates Innovative Ability in Scientific Research of Huaqiao University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any study with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 360 kb)
Rights and permissions
About this article
Cite this article
Ke, C., Zhao, C., Rensing, C. et al. Characterization of recombinant E. coli expressing arsR from Rhodopseudomonas palustris CGA009 that displays highly selective arsenic adsorption. Appl Microbiol Biotechnol 102, 6247–6255 (2018). https://doi.org/10.1007/s00253-018-9080-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9080-8