Abstract
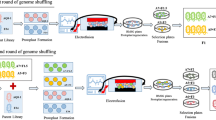
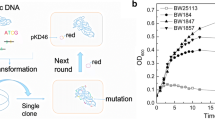
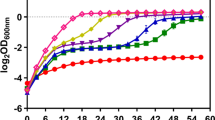
Furfural-tolerant strain is essential for the fermentative production of biofuels or chemicals from lignocellulosic biomass. In this study, Zymomonas mobilis CP4 was for the first time subjected to error-prone PCR-based whole genome shuffling, and the resulting mutants F211 and F27 that could tolerate 3 g/L furfural were obtained. The mutant F211 under various furfural stress conditions could rapidly grow when the furfural concentration reduced to 1 g/L. Meanwhile, the two mutants also showed higher tolerance to high concentration of glucose than the control strain CP4. Genome resequencing revealed that the F211 and F27 had 12 and 13 single-nucleotide polymorphisms. The activity assay demonstrated that the activity of NADH-dependent furfural reductase in mutant F211 and CP4 was all increased under furfural stress, and the activity peaked earlier in mutant than in control. Also, furfural level in the culture of F211 was also more rapidly decreased. These indicate that the increase in furfural tolerance of the mutants may be resulted from the enhanced NADH-dependent furfural reductase activity during early log phase, which could lead to an accelerated furfural detoxification process in mutants. In all, we obtained Z. mobilis mutants with enhanced furfural and high concentration of glucose tolerance, and provided valuable clues for the mechanism of furfural tolerance and strain development.




Similar content being viewed by others
References
Agrawal M, Chen RR (2011) Discovery and characterization of a xylose reductase from Zymomonas mobilis ZM4. Biotechnol Lett 33(11):2127–2133. https://doi.org/10.1007/s10529-011-0677-6
Agrawal M, Mao Z, Chen RR (2011) Adaptation yields a highly efficient xylose-fermenting Zymomonas mobilis strain. Biotechnol Bioeng 108(4):777–785. https://doi.org/10.1002/bit.23021
Ajit A, Sulaiman AZ, Chisti Y (2017) Production of bioethanol by Zymomonas mobilis in high-gravity extractive fermentations. Food Bioprod Process 102:123–135. https://doi.org/10.1016/j.fbp.2016.12.006
Almeida JR, Modig T, Petersson A, Hähn-Hägerdal B, Lidén G, Gorwa-Grauslund MF (2007) Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82(4):340–349. https://doi.org/10.1002/jctb.1676
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72(1–2):248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Bui LM, Lee JY, Geraldi A, Rahman Z, Lee JH, Kim SC (2015) Improved n-butanol tolerance in Escherichia coli by controlling membrane related functions. J Biotechnol 204:33–44. https://doi.org/10.1016/j.jbiotec.2015.03.025
Carey VC, Ingram LO (1983) Lipid composition of Zymomonas mobilis: effects of ethanol and glucose. J Bacteriol 1291–1300
Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R (2002) Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol 184(3):754–759. https://doi.org/10.1128/JB.184.3.754-759.2002
Emanuelsson EA, Baptista II, Mantalaris A, Livingston AG (2005) Strain stability in biological systems treating recalcitrant organic compounds. Biotechnol Bioeng 92(7):843–849. https://doi.org/10.1002/bit.20620
Fillat MF, Borrias WE, Weisbeek PJ (1991) Isolation and overexpression in Escherichia coli of the flavodoxin gene from Anabaena PCC 7119. Biochem J 280(1):187–191. https://doi.org/10.1042/bj2800187
Franden MA, Pilath HM, Mohagheghi A, Pienkos PT, Zhang M (2013) Inhibition of growth of Zymomonas mobilis by model compounds found in lignocellulosic hydrolysates. Biotechnol Biofuels 6(1):99. https://doi.org/10.1186/1754-6834-6-99
Groth AC, Calos MP (2004) Phage integrases: biology and applications. J Mol Biol 335(3):667–678. https://doi.org/10.1016/j.jmb.2003.09.082
Gu H, Zhang J, Bao J (2015) High tolerance and physiological mechanism of Zymomonas mobilis to phenolic inhibitors in ethanol fermentation of corncob residue. Biotechnol Bioeng 112(9):1770–1782. https://doi.org/10.1002/bit.25603
Hadar Y (2013) Sources for lignocellulosic raw materials for the production of ethanol. In: Lignocellulose Conversion. Springer Berlin, Heidelberg, pp 21–38. https://doi.org/10.1007/978-3-642-37861-4_2
Haruta S, Cui Z, Huang Z, Li M, Ishii M, Igarashi Y (2002) Construction of a stable microbial community with high cellulose-degradation ability. Appl Microbiol Biotechnol 59(4):529–534
He MX, Wu B, Shui ZX, Hu QC, Wang WG, Tan FR, Tang XY, Zhu QL, Pan K, Li Q (2012) Transcriptome profiling of Zymomonas mobilis under furfural stress. Appl Microbiol Biotechnol 95(1):189–199. https://doi.org/10.1007/s00253-012-4155-4
He MX, Wu B, Qin H, Ruan ZY, Tan FR, Wang JL, Shui ZX, Dai LC, Zhu QL, Pan K (2014) Zymomonas mobilis: a novel platform for future biorefineries. Biotechnol Biofuels 7(1):101. https://doi.org/10.1186/1754-6834-7-101
Jeon YJ, Xun Z, Su P, Rogers PL (2012) Genome-wide transcriptomic analysis of a flocculent strain of Zymomonas mobilis. Appl Microbiol Biotechnol 93(6):2513–2518. https://doi.org/10.1007/s00253-012-3948-9
Kádár Z, Maltha SF, Szengyel Z, Réczey K, De Laat W (2007) Ethanol fermentation of various pretreated and hydrolyzed substrates at low initial pH. Appl Biochem Biotech 137(1–12):847–858
Kerr AL, Jeon YJ, Svenson CJ, Rogers PL, Neilan BA (2011) DNA restriction-modification systems in the ethanologen, Zymomonas mobilis ZM4. Appl Microbiol Biotechnol 89(3):761–769. https://doi.org/10.1007/s00253-010-2936-1
Kim HK, Harshey RM (2016) A diguanylate cyclase acts as a cell division inhibitor in a two-step response to reductive and envelope stresses. MBio 7(4):e00822–e00816
Kim MS, Lei XG (2008) Enhancing thermostability of Escherichia coli phytase AppA2 by error-prone PCR. Appl Microbiol Biotechnol 79(1):69–75. https://doi.org/10.1007/s00253-008-1412-7
Liao Y, Zeng M, Wu ZF, Chen H, Wang HN, Wu Q, Shan Z, Han XY (2012) Improving phytase enzyme activity in a recombinant phyA mutant phytase from Aspergillus niger N25 by error-prone PCR. Appl Biochem Biotechnol 166(3):549–562. https://doi.org/10.1007/s12010-011-9447-0
Lin Y, Tanaka S (2006) Ethanol fermentation from biomass resources: current state and prospects. Appl Microbiol Biotechnol 69(6):627–642. https://doi.org/10.1007/s00253-005-0229-x
Liu ZL, Moon J (2009) A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446(1):1–10. https://doi.org/10.1016/j.gene.2009.06.018
Liu ZL, Moon J, Andersh BJ, Slininger PJ, Weber S (2008) Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81(4):743–753. https://doi.org/10.1007/s00253-008-1702-0
Luhe AL, Tan L, Wu J, Hua Z (2011) Increase of ethanol tolerance of Saccharomyces cerevisiae by error-prone whole genome amplification. Biotechnol Lett 33(5):1007–1011. https://doi.org/10.1007/s10529-011-0518-7
Ma Y, Dong H, Zou S, Hong J, Zhang M (2012) Comparison of glucose/xylose co-fermentation by recombinant Zymomonas mobilis under different genetic and environmental conditions. Biotechnol Lett 34(7):1297–1304. https://doi.org/10.1007/s10529-012-0897-4
Martínez-Patiño JC, Ruiz E, Romero I, Cara C, López-Linares JC, Castro E (2017) Combined acid/alkaline-peroxide pretreatment of olive tree biomass for bioethanol production. Bioresour Technol 239:326–335. https://doi.org/10.1016/j.biortech.2017.04.102
Mills TY, Sandoval NR, Gill RT (2009) Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol Biofuels 2(1):26. https://doi.org/10.1186/1754-6834-2-26
Moon J, Liu ZL (2015) Direct enzyme assay evidence confirms aldehyde reductase function of Ydr541cp and Ygl039wp from Saccharomyces cerevisiae. Yeast 32(4):399–407. https://doi.org/10.1002/yea.3067
Moreno AD, Ibarra D, Alvira P, Tomás-Pejó E, Ballesteros M (2015) A review of biological delignification and detoxification methods for lignocellulosic bioethanol production. Crit Rev Biotechnol 35(3):342–354. https://doi.org/10.3109/07388551.2013.878896
Puligundla P, Smogrovicova D, Obulam VS, Ko S (2011) Very high gravity (VHG) ethanolic brewing and fermentation: a research update. J Ind Microbiol Biotechnol 38(9):1133–1144. https://doi.org/10.1007/s10295-011-0999-3
Romling U, Galperin MY, Gomelsky M (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77(1):1–52. https://doi.org/10.1128/MMBR.00043-12
Shi D-J, Wang C-L, Wang K-M (2009) Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 36(1):139–147. https://doi.org/10.1007/s10295-008-0481-z
Shui ZX, Qin H, Wu B, Ruan ZY, Wang LS, Tan FR, Wang JL, Tang XY, Dai LC, Hu GQ (2015) Adaptive laboratory evolution of ethanologenic Zymomonas mobilis strain tolerant to furfural and acetic acid inhibitors. Appl Microbiol Biotechnol 99(13):5739–5748. https://doi.org/10.1007/s00253-015-6616-z
Tan FR, Dai LC, Wu B, Qin H, Shui ZX, Wang JL, Zhu QL, Hu QC, Ruan ZY, He MX (2015) Improving furfural tolerance of Zymomonas mobilis by rewiring a sigma factor RpoD protein. Appl Microbiol Biotechnol 99(12):5363–5371. https://doi.org/10.1007/s00253-015-6577-2
Todhanakasem T, Sangsutthiseree A, Areerat K, Young GM, Thanonkeo P (2014) Biofilm production by Zymomonas mobilis enhances ethanol production and tolerance to toxic inhibitors from rice bran hydrolysate. New Biotechnol 31(5):451–459. https://doi.org/10.1016/j.nbt.2014.06.002
Todhanakasem T, Narkmit T, Areerat K, Thanonkeo P (2015) Fermentation of rice bran hydrolysate to ethanol using Zymomonas mobilis biofilm immobilization on DEAE-cellulose. Electron J Biotechnol 18(3):196–201. https://doi.org/10.1016/j.ejbt.2015.03.007
Wang X, Miller EN, Yomano LP, Zhang X, Shanmugam KT, Ingram LO (2011) Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl Environ Microbiol 77(15):5132–5140. https://doi.org/10.1128/AEM.05008-11
Wang X, Ma M, Liu ZL, Xiang Q, Li X, Liu N, Zhang X (2016a) GRE2 from Scheffersomyces stipitis as an aldehyde reductase contributes tolerance to aldehyde inhibitors derived from lignocellulosic biomass. Appl Microbiol Biotechnol 100(15):6671–6682. https://doi.org/10.1007/s00253-016-7445-4
Wang H, Cao S, Wang WT, Wang KT, Jia X (2016b) Very high gravity ethanol and fatty acid production of Zymomonas mobilis without amino acid and vitamin. J Ind Microbiol Biotechnol 43(6):861–871. https://doi.org/10.1007/s10295-016-1761-7
Wang X, Gao Q, Bao J (2017) Enhancement of furan aldehydes conversion in Zymomonas mobilis by elevating dehydrogenase activity and cofactor regeneration. Biotechnol Biofuels 10(1):24. https://doi.org/10.1186/s13068-017-0714-3
Yang S, Pelletier DA, Lu TYS, Brown SD (2010) The Zymomonas mobilis regulator hfq contributes to tolerance against multiple lignocellulosic pretreatment inhibitors. BMC Microbiol 10(1):135. https://doi.org/10.1186/1471-2180-10-135
Yang S, Franden MA, Brown SD, Chou Y-C, Pienkos PT, Zhang M (2014) Insights into acetate toxicity in Zymomonas mobilis 8b using different substrates. Biotechnol Biofuels 7(1):140. https://doi.org/10.1186/s13068-014-0140-8
Ye L, Zhao H, Li Z, Wu JC (2013) Improved acid tolerance of Lactobacillus pentosus by error-prone whole genome amplification. Bioresour Technol 135(10):459–463. https://doi.org/10.1016/j.biortech.2012.10.042
Yi X, Gu H, Gao Q, Liu ZL, Bao J (2015) Transcriptome analysis of Zymomonas mobilis ZM4 reveals mechanisms of tolerance and detoxification of phenolic aldehyde inhibitors from lignocellulose pretreatment. Biotechnol Biofuels 8(1):153. https://doi.org/10.1186/s13068-015-0333-9
You Y, Liu S, Wu B, Wang YW, Zhu QL, Qin H, Tan FR, Ruan ZY, Ma KD, Dai LC (2017) Bio-ethanol production by Zymomonas mobilis using pretreated dairy manure as a carbon and nitrogen source. RSC Adv 7(7):3768–3779. https://doi.org/10.1039/C6RA26288K
Zhang K, Shao HH, Cao QH, He MX, Wu B, Feng H (2015) Transcriptional analysis of adaptation to high glucose concentrations in Zymomonas mobilis. Appl Microbiol Biotechnol 99(4):2009–2022. https://doi.org/10.1007/s00253-014-6342-y
Acknowledgements
We thank Dr. Jie Bao for providing plasmid pHW20a.
Funding
This research was supported by the National Natural Science Foundation of China (NSFC30900033, NSFC21376174) and the Tianjin Science and Technology Council (13JCYBJC40700).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 1048 kb)
Rights and permissions
About this article
Cite this article
Huang, S., Xue, T., Wang, Z. et al. Furfural-tolerant Zymomonas mobilis derived from error-prone PCR-based whole genome shuffling and their tolerant mechanism. Appl Microbiol Biotechnol 102, 3337–3347 (2018). https://doi.org/10.1007/s00253-018-8817-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-8817-8