Abstract
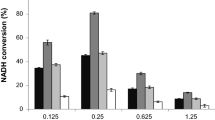
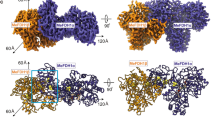
Formate dehydrogenases (FDHs) are considered particularly useful enzymes in biocatalysis when the regeneration of the cofactor NAD(P)H is required, that is, in chiral synthesis with dehydrogenases. Their utilization is however limited to the recycling of NAD+, since all (apart one) of the FDHs characterized so far are strictly specific for this cofactor, and this is a major drawback for their otherwise wide applicability. Despite the many attempts performed to modify cofactor specificity by protein engineering different NAD+-dependent FDHs, in the general practice, glucose or phosphite dehydrogenases are chosen for the recycling of NADP+. We report on the functional and structural characterization of a new FDH, GraFDH, identified by mining the genome of the extremophile prokaryote Granulicella mallensis MP5ACTX8. The new enzyme displays a valuable stability in the presence of many organic cosolvents as well as double cofactor specificity, with NADP+ preferred over NAD+ at acidic pH values, at which it also shows the highest stability. The quite low affinities for both cofactors as well as for the substrate formate indicate, however, that the native enzyme requires optimization to be applied as biocatalytic tool. We also determined the crystal structure of GraFDH both as apoprotein and as holoprotein, either in complex with NAD+ or NADP+. Noticeably, the latter represents the first structure of an FDH enzyme in complex with NADP+. This fine picture of the structural determinants involved in cofactor selectivity will possibly boost protein engineering of the new enzyme or other homolog FDHs in view of their biocatalytic exploitation for NADP+ recycling.





Similar content being viewed by others
References
Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L, Kapral GJ, Grosse-Kunstleve RW (2010) PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221
Alekseeva A, Savin S, Tishkov V (2011) NAD-dependent formate dehydrogenase from plants. Acta Nat 3:38–54
Andreadeli A, Platis D, Tishkov V, Popov V, Labrou NE (2008) Structure‐guided alteration of coenzyme specificity of formate dehydrogenase by saturation mutagenesis to enable efficient utilization of NADP. FEBS J 275:3859–3869
Bommarius AS, Schwarm M, Stingl K, Kottenhahn M, Huthmacher K, Drauz K (1995) Synthesis and use of enantiomerically pure tert-leucine. Tetrahedron Asymmetry 6:2851–2888
Emsley P, Lohkamp B, Scott W, Cowtan K (2010) Features and development of coot. Acta Crystallogr D Biol Crystallogr 66:486–501
Ericsson UB, Hallberg BM, DeTitta GT, Dekker N, Nordlund P (2006) Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem 357:289–298
Evans P (2005) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82
Gul-Karaguler N, Sessions RB, Clarke AR, Holbrook JJ (2001) A single mutation in the NAD-specific formate dehydrogenase from Candida methylica allows the enzyme to use NADP. Biotechnol Lett 23:283–287
Hatrongjit R, Packdibamrung K (2010) A novel NADP+-dependent formate dehydrogenase from Burkholderia stabilis 15516: screening, purification and characterization. Enzym Microb Technol 46:557–561
Hoelsch K, Sührer I, Heusel M, Weuster-Botz D (2013) Engineering of formate dehydrogenase: synergistic effect of mutations affecting cofactor specificity and chemical stability. Appl Microbiol Biotechnol 97:2473–2481
Ihara M, Kawano Y, Urano M, Okabe A (2013) Light driven CO2 fixation by using cyanobacterial photosystem I and NADPH-dependent formate dehydrogenase. PLoS One 8: e71581
Johannes TW, Woodyer RD, Zhao H (2007) Efficient regeneration of NADPH using an engineered phosphite dehydrogenase. Biotechnol Bioeng 96:18–26
Kaswurm V, Hecke WV, Kulbe KD, Ludwig R (2013) Guidelines for the application of NAD(P)H regenerating glucose dehydrogenase in synthetic processes. Adv Synth Catal 355:1709–1714
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797
Lamzin VS, Dauter Z, Popov VO, Harutyunyan EH, Wilson KS (1994) High resolution structures of holo and apo formate dehydrogenase. J Mol Biol 236:759–785
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Laskowski RA, Watson JD, Thornton JM (2005) ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res 33:W89–W93
Lavinder JJ, Hari SB, Sullivan BJ, Magliery TJ (2009) High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J Am Chem Soc 131:3794–3795
Männistö MK, Rawat S, Starovoytov V, Häggblom MM (2012) Granulicella arctica sp. nov., Granulicella mallensis sp. nov., Granulicella tundricola sp. nov. and Granulicella sapmiensis sp. nov., novel acidobacteria from tundra soil. Int J Syst Evol Microbiol 62:2097–2106
Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367
Popov VO, Lamzin VS (1994) NAD+-dependent formate dehydrogenase. Biochem J 301(Pt 3):625–643
Rawat SR, Männistö MK, Starovoytov V, Goodwin L, Nolan M, Hauser LJ, Land M, Davenport KW, Woyke T, Häggblom MM (2013) Complete genome sequence of Granulicella mallensis type strain MP5ACTX8T, an acidobacterium from tundra soil. Stand Genomic Sci 9:71
Relyea HA, Vrtis JM, Woodyer R, Rimkus SA, van der Donk WA (2005) Inhibition and pH dependence of phosphite dehydrogenase. Biochemistry 44:6640–6649
Sadykhov E, Serov A, Voinova N, Uglanova S, Petrov A, Alekseeva A, Kleimenov SY, Popov V, Tishkov V (2006) A comparative study of the thermal stability of formate dehydrogenases from microorganisms and plants. Appl Biochem Microbiol 42:236–240
Schirwitz K, Schmidt A, Lamzin VS (2007) High-resolution structures of formate dehydrogenase from Candida boidinii. Protein Sci 16:1146–1156
Serov A, Popova A, Fedorchuk V, Tishkov V (2002) Engineering of coenzyme specificity of formate dehydrogenase from Saccharomyces cerevisiae. Biochem J 367:841–847
Shabalin I, Filippova E, Polyakov K, Sadykhov E, Safonova T, Tikhonova T, Tishkov V, Popov V (2009) Structures of the apo and holo forms of formate dehydrogenase from the bacterium Moraxella sp. C-1: towards understanding the mechanism of the closure of the interdomain cleft. Acta Crystallogr D Biol Crystallogr 65:1315–1325
Tishkov V, Popov V (2004) Catalytic mechanism and application of formate dehydrogenase. Biochem Mosc 69:1252–1267
Tishkov VI, Popov VO (2006) Protein engineering of formate dehydrogenase. Biomol Eng 23:89–110
Weckbecker A, Groger H, Hummel V (2010) Regeneration of nicotinamide coenzymes: principles and applications for the synthesis of chiral compounds. Adv Biochem Eng Biotechnol 120:195–242
Wong C, Drueckhammer DG, Sweers HM (1985) Enzymatic vs. fermentative synthesis: thermostable glucose dehydrogenase catalyzed regeneration of NAD(P)H for use in enzymatic synthesis. J Am Chem Soc 107:4028–4031
Woodyer R, van der Donk WA, Zhao H (2003) Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design. Biochemistry 42:11604–11614
Wu JT, Wu LH, Knight JA (1986) Stability of NADPH: effect of various factors on the kinetics of degradation. Clin Chem 32:314–319
Wu W, Zhu D, Hua L (2009a) Site-saturation mutagenesis of formate dehydrogenase from Candida boidinii creating effective NADP+-dependent FDH enzymes. J Mol Catal B 61:157–161
Wu X, Wang L, Wang S, Chen Y (2009b) Stereoselective introduction of two chiral centers by a single diketoreductase: an efficient biocatalytic route for the synthesis of statin side chains. Amino Acids 39:305–308
Acknowledgments
This work has been financed by an Assegno di Ricerca (CPDR095797/09) from the University of Padova (Italy) to S. Fogal and by a grant from Fabbrica Italiana Sintetici-F.I.S. S.p.A. (Montecchio Maggiore, Vicenza; Italy) to E. Bergantino.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Stefano Fogal and Elisa Beneventi contributed equally to the work.
Electronic supplementary material
ESM 1
(PDF 663 kb)
Rights and permissions
About this article
Cite this article
Fogal, S., Beneventi, E., Cendron, L. et al. Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8. Appl Microbiol Biotechnol 99, 9541–9554 (2015). https://doi.org/10.1007/s00253-015-6695-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6695-x