Abstract
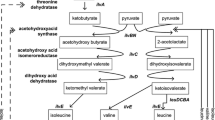
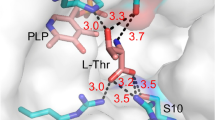
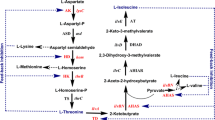
Biosynthetic threonine deaminase (TD) is a key enzyme for the synthesis of isoleucine which is allosterically inhibited and activated by Ile and Val, respectively. The binding sites of Ile and Val and the mechanism of their regulations in TD are not clear, but essential for a rational design of efficient productive strain(s) for Ile and related amino acids. In this study, structure-based computational approach and site-directed mutagenesis were combined to identify the potential binding sites of Ile and Val in Escherichia coli TD. Our results demonstrated that each regulatory domain of the TD monomer possesses two nonequivalent effector-binding sites. The residues R362, E442, G445, A446, Y369, I460, and S461 only interact with Ile while E347, G350, and F352 are involved not only in the Ile binding but also in the Val binding. By further considering enzyme kinetic data, we propose a concentration-dependent mechanism of the allosteric regulation of TD by Ile and Val. For the construction of Ile overproducing strain, a novel TD mutant with double mutation of F352A/R362F was also created, which showed both higher activity and much stronger resistance to Ile inhibition comparing to those of wild-type enzyme. Overexpression of this mutant TD in E. coli JW3591 significantly increased the production of ketobutyrate and Ile in comparison to the reference strains overexpressing wild-type TD or the catabolic threonine deaminase (TdcB). This work builds a solid basis for the reengineering of TD and related microorganisms for Ile production.








Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Amann E, Ochs B, Abel KJ (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315
Anantharaman V, Koonin EV, Aravind L (2001) Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol 307:1271–1292. doi:10.1006/jmbi.2001.4508
Aravind L, Koonin EV (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol 287:1023–1040
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89. doi:10.1038/nature06450
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi:10.1038/msb4100050
Chen Z, Meyer W, Rappert S, Sun J, Zeng AP (2011a) Coevolutionary analysis enabled rational deregulation of allosteric enzyme inhibition in Corynebacterium glutamicum for lysine production. Appl Environ Microbiol 77:4352–4360. doi:10.1128/AEM.02912-10
Chen Z, Rappert S, Sun J, Zeng AP (2011b) Integrating molecular dynamics and co-evolutionary analysis for reliable target prediction and deregulation of the allosteric inhibition of aspartokinase for amino acid production. J Biotechnol 154:248–254. doi:10.1016/j.jbiotec.2011.05.005
Chipman DM, Shaanan B (2001) The ACT domain family. Curr Opin Struct Biol 11:694–700
Curien G, Biou V, Mas-Droux C, Robert-Genthon M, Ferrer JL, Dumas R (2008) Amino acid biosynthesis: new architectures in allosteric enzymes. Plant Physiol Biochem 46:325–339. doi:10.1016/j.plaphy.2007.12.006
Davis L (1965) A spectrophotometric method for the assay of threonine dehydratase. Anal Biochem 12:36–40
Decedue CJ, Hofler JG, Burns RO (1975) Threonine deaminase from Salmonella typhimurium. Relationship between regulatory sites. J Biol Chem 250:1563–1570
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi:10.1093/nar/gkh340
Eisenstein E (1991) Cloning, expression, purification, and characterization of biosynthetic threonine deaminase from Escherichia coli. J Biol Chem 266:5801–5807
Eisenstein E, Yu HD, Schwarz FP (1994) Cooperative binding of the feedback modifiers isoleucine and valine to biosynthetic threonine deaminase from Escherichia coli. J Biol Chem 269:29423–29429
Eisenstein E, Yu HD, Fisher KE, Iacuzio DA, Ducote KR, Schwarz FP (1995) An expanded two-state model accounts for homotropic cooperativity in biosynthetic threonine deaminase from Escherichia coli. Biochemistry 34:9403–9412
Feldman DA, Datta P (1975) Catabolite inactivation of biodegradative threonine dehydratase of Escherichia coli. Biochemistry 14:1760–1767
Gallagher DT, Gilliland GL, Xiao G, Zondlo J, Fisher KE, Chinchilla D, Eisenstein E (1998) Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure 6:465–475
Gallagher DT, Chinchilla D, Lau H, Eisenstein E (2004) Local and global control mechanisms in allosteric threonine deaminase. Methods Enzymol 380:85–106. doi:10.1016/S0076-6879(04)80004-1
Grant GA (2006) The ACT domain: a small molecule binding domain and its role as a common regulatory element. J Biol Chem 281:33825–33829. doi:10.1074/jbc.R600024200
Gruys KJ, Mitsky TA, Kishore GM, Slater SC, Padgette SR, Stark DM, Hinchee MA, Clemente TE, Connor-Ward D, Fedele MJ, Fry JE, Howe AR, Rozman RJ, Hinchee MAW, Connor-Ward DV (2007) Modified threonine deaminase. US Patent 7:192,753
Guillouet S, Rodal AA, An G, Lessard PA, Sinskey AJ (1999) Expression of the Escherichia coli catabolic threonine dehydratase in Corynebacterium glutamicum and its effect on isoleucine production. Appl Environ Microbiol 65:3100–3107
Guillouet S, Rodal AA, An GH, Gorret N, Lessard PA, Sinskey AJ (2001) Metabolic redirection of carbon flow toward isoleucine by expressing a catabolic threonine dehydratase in a threonine-overproducing Corynebacterium glutamicum. Appl Microbiol Biotechnol 57:667–673
Halabi N, Rivoire O, Leibler S, Ranganathan R (2009) Protein sectors: evolutionary units of three-dimensional structure. Cell 138:774–786. doi:10.1016/j.cell.2009.07.038
Hashiguchi K, Takesada H, Suzuki E, Matsui H (1999) Construction of an L-isoleucine overproducing strain of Escherichia coli K-12. Biosci Biotechnol Biochem 63:672–679
Heinrikson RL, Meredith SC (1984) Amino acid analysis by reverse-phase high-performance liquid chromatography: precolumn derivatization with phenylisothiocyanate. Anal Biochem 136:65–74
Kaplun A, Vyazmensky M, Zherdev Y, Belenky I, Slutzker A, Mendel S, Barak Z, Chipman DM, Shaanan B (2006) Structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from Escherichia coli. J Mol Biol 357:951–963. doi:10.1016/j.jmb.2005.12.077
Koerner K, Rahimi-Laridjani I, Grimminger H (1975) Purification of biosynthetic threonine deaminase from Escherichia coli. Biochim Biophys Acta 397:220–230
Kotaka M, Ren J, Lockyer M, Hawkins AR, Stammers DK (2006) Structures of R- and T-state Escherichia coli aspartokinase III. Mechanisms of the allosteric transition and inhibition by lysine. J Biol Chem 281:31544–31552. doi:10.1074/jbc.M605886200
Mas-Droux C, Curien G, Robert-Genthon M, Laurencin M, Ferrer JL, Dumas R (2006) A novel organization of ACT domains in allosteric enzymes revealed by the crystal structure of Arabidopsis aspartate kinase. Plant Cell 18:1681–1692. doi:10.1105/tpc.105.040451
Monod J, Wyman J, Changeux JP (1965) On the nature of allosteric transitions: a plausible model. J Mol Biol 12:88–118
Park JH, Lee SY (2010) Fermentative production of branched chain amino acids: a focus on metabolic engineering. Appl Microbiol Biotechnol 85:491–506. doi:10.1007/s00253-009-2307-y
Schuller DJ, Grant GA, Banaszak LJ (1995) The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat Struct Biol 2:69–76
Shatsky M, Nussinov R, Wolfson HJ (2004) A method for simultaneous alignment of multiple protein structures. Proteins 56:143–156. doi:10.1002/prot.10628
Shizuta Y, Kurosawa A, Inoue K, Tanabe T, Hayaishi O (1973) Regulation of biodegradative threonine deaminase. I. Allosteric inhibition of the enzyme by a reaction product and its reversal by adenosin 5′-monophosphate. J Biol Chem 248:512–520
Shulman A, Zalyapin E, Vyazmensky M, Yifrach O, Barak Z, Chipman DM (2008) Allosteric regulation of Bacillus subtilis threonine deaminase, a biosynthetic threonine deaminase with a single regulatory domain. Biochemistry 47:11783–11792. doi:10.1021/bi800901n
Umbarger HE (1956) Evidence for a negative-feedback mechanism in the biosynthesis of isoleucine. Science 123:848
Wessel PM, Graciet E, Douce R, Dumas R (2000) Evidence for two distinct effector-binding sites in threonine deaminase by site-directed mutagenesis, kinetic, and binding experiments. Biochemistry 39:15136–15143
Yan Y, Liao JC (2009) Engineering metabolic systems for production of advanced fuels. J Ind Microbiol Biotechnol 36:471–479. doi:10.1007/s10295-009-0532-0
Yoshida A, Tomita T, Kurihara T, Fushinobu S, Kuzuyama T, Nishiyama M (2007) Structural Insight into concerted inhibition of alpha 2 beta 2-type aspartate kinase from Corynebacterium glutamicum. J Mol Biol 368:521–536. doi:10.1016/j.jmb.2007.02.017
Acknowledgments
The authors LC, PZ, and JS gratefully thank the financial support from the 973 key basic research program with the project 2011CBA00804 and the Chinese Academy of Sciences with the project KSCX2-EW-G-14-1. ZC and APZ were supported by the German Research Foundation (DFG) through the project ZE 542/6-1 and the Hamburg Excellence Initiative project SynBio.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Lin Chen and Zhen Chen equally contributed to this work.
Rights and permissions
About this article
Cite this article
Chen, L., Chen, Z., Zheng, P. et al. Study and reengineering of the binding sites and allosteric regulation of biosynthetic threonine deaminase by isoleucine and valine in Escherichia coli . Appl Microbiol Biotechnol 97, 2939–2949 (2013). https://doi.org/10.1007/s00253-012-4176-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4176-z