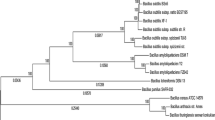
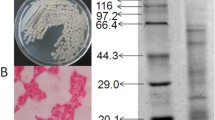
Abstract
Chemotaxis is a process in which bacteria sense their chemical environment and move towards more favorable conditions. Since plant colonization by bacteria is a multifaceted process which requires a response to the complex chemical environment, a finely tuned and sensitive chemotaxis system is needed. Members of the Bacillus subtilis group including Bacillus amyloliquefaciens are industrially important, for example, as bio-pesticides. The group exhibits plant growth-promoting characteristics, with different specificity towards certain host plants. Therefore, we hypothesize that while the principal molecular mechanisms of bacterial chemotaxis may be conserved, the bacterial chemotaxis system may need an evolutionary tweaking to adapt it to specific requirements, particularly in the process of evolution of free-living soil organisms, towards plant colonization behaviour. To date, almost nothing is known about what parts of the chemotaxis proteins are subjected to positive amino acid substitutions, involved in adjusting the chemotaxis system of bacteria during speciation. In this novel study, positively selected and purified sites of chemotaxis proteins were calculated, and these residues were mapped onto homology models that were built for the chemotaxis proteins, in an attempt to understand the spatial evolution of the chemotaxis proteins. Various positively selected amino acids were identified in semi-conserved regions of the proteins away from the known active sites.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Borriss R, Chen X, Rueckert C, Blom J, Becker A, Baumgarth B, Fan B, Pukall R, Schumann P, Spröer C, Junge H, Vater J, Pühler A, Klenk HP (2010) Relationship of Bacillus amyloliquefaciens clades associated with strains DSM7T and FZB42: a proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on their discriminating complete genome sequences. Int J Syst Evol Microbiol. doi:10.1099/ijs.0.023267-0
Buchan A, Crombie B, Alexandre GM (2010) Temporal dynamics and genetic diversity of chemotactic-competent microbial populations in the rhizosphere. Environ Microbiol 12(12):3171–3184
Bunn MW, Ordal GW (2003) Transmembrane organization of the Bacillus subtilis chemoreceptor McpB deduced by cysteine disulfide crosslinking. J Mol Biol 331:941–949
Chao X, Muff TJ, Park S, Zhang S, Pollard AM, Ordal GW, Bilwes AM, Crane BR (2006) A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 124:561–571
Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva O, Junge H, Voigt B, Jungblut PR, Vater J, Süssmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R (2007) Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotech 25:1007–1014
Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R (2009) Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol 140:27–37
Choudhary DK, Johri BN (2009) Interactions of Bacillus spp. and plants—with special reference to induced. Microbiol Res 164(5):493–513
Chun J, Bae KS (2000) Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie van Leeuwenhoek 78:123–127
Djordjevic S, Stock AM (1997) Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure 5:545–558
Djordjevic S, Goudreau PN, Xu Q, Stock AM, West AH (1998) Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc Natl Acad Sci USA 95:1381–1386
Doron-Faigenboim A, Pupko T (2007) A combined empirical and mechanistic codon model. Mol Biol Evol 24:388–397
Doron-Faigenboim A, Stern A, Mayrose I, Bacharach E, Pupko T (2005) Selecton: a server for detecting evolutionary forces at a single amino-acid site. Bioinformatics 21:2101–2103
Edgar R (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Falke JJ, Hazelbauer GL (2001) Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci 26:257–265
Felsenstein J, Churchill GA (1996) A Hidden Markov Model approach to variation among sites in rate of evolution. Mol Biol Evol 13:93–104
Fritze D (2004) Taxonomy of the genus Bacillus and related genera: the aerobic endospore-forming bacteria. Phytopathology 94:1245–1248
Garrity LF, Ordal GW (1995) Chemotaxis in Bacillus subtilis: how bacteria monitor environmental signals. Pharmacol Ther 68:87–104
Garrity LF, Schiel SL, Merrill R, Reizer J, Saier MH Jr, Ordal GW (1998) Unique regulation of carbohydrate chemotaxis in Bacillus subtilis by the phosphoenolpyruvate-dependent phosphotransferase system and the methyl-accepting chemotaxis protein McpC. J Bacteriol 180:4475–4480
Goldman DJ, Ordal GW (1984) In vitro methylation and demethylation of methyl-accepting chemotaxis proteins in Bacillus subtilis. Biochemistry 23:2600–2606
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hamer R, Chen P, Armitage J, Reinert G, Deane C (2010) Deciphering chemotaxis pathways using cross species comparisons. BMC Systems Biology 4:3
Hernández-Sánchez C, Mansilla A, de Pablo F, Zardoya R (2008) Evolution of the insulin receptor family and receptor isoform expression in vertebrates. Mol Biol Evol 25:1043–1053
Kirby JR, Niewold TB, Maloy S, Ordal GW (2000) CheB is required for behavioural responses to negative stimuli during chemotaxis in Bacillus subtilis. Mol Microbiol 35:44–57
Kirby JR, Kristich CJ, Saulmon M, Zimmer MA, Garrity L, Zhulin IB, Ordal G (2001) CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol Microbiol 42:573–585
Kristich CJ, Ordal GW (2002) Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J Biol Chem 277:25356–25362
Kristich CJ, Ordal GW (2004) Analysis of chimeric chemoreceptors in Bacillus subtilis reveals a role for CheD in the function of the McpC HAMP domain. J Bacteriol 186:5950–5955
Kristich CJ, Glekas GD, Ordal GW (2003) The conserved cytoplasmic module of the transmembrane chemoreceptor McpC mediates carbohydrate chemotaxis in Bacillus subtilis. Mol Microbiol 47:1353–1366
Lupas A, Stock J (1989) Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem 264:17337–17342
Lux R, Munasinghe VRN, Castellano F, Lengeler JW, Corrie JET, Khan S (1999) Elucidation of a PTS-carbohydrate chemotactic signal pathway in Escherichia coli using a time-resolved behavioral assay. Mol Biol Cell 10:1133–1146
Manson MD, Armitage JP, Hoch JA, Macnab RM (1998) Bacterial locomotion and signal transduction. J Bacteriol 180:1009–1022
Mowbray SL, Sandgren MOJ (1998) Chemotaxis receptors: a progress report on structure and function. J Struct Biol 124:257–275
Muff TJ, Ordal GW (2007) The CheC phosphatase regulates chemotactic adaptation through CheD. J Biol Chem 282:34120–34128
Park S, Chao X, Gonzalez-Bonet G, Beel BD, Bilwes AM, Crane BR (2004a) Structure and function of an unusual family of protein phosphatases: the bacterial chemotaxis proteins CheC and CheX. Mol Cell 16:563–574
Park S, Beel BD, Simon MI, Bilwes AM, Crane BR (2004b) In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc Natl Acad Sci USA 101:11646–11651
Park S, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR (2006) Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol 13:400–407
Pawlowski M, Gajda M, Matlak R, Bujnicki J (2008) MetaMQAP: a meta-server for the quality assessment of protein models. BMC Bioinformatics 9:403
Pazy Y, Motaleb MA, Guarnieri MT, Charon NW, Zhao R, Silversmith RE (2010) Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate. Proc Natl Acad Sci USA 107:1924–1929
Pei J, Kim B, Grishin NV (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucl Acids Res 36:2295–2300
Rao CV, Kirby JR, Arkin AP (2004) Design and diversity in bacterial chemotaxis: a comparative study in Escherichia coli and Bacillus subtilis. PLoS Biol 2:e49
Rao CV, Glekas GD, Ordal GW (2008) The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol 16:480–487
Reva ON, Dixelius C, Meijer J, Priest FG (2004) Taxonomic characterization and plant colonizing abilities of some bacteria related to Bacillus amyloliquefaciens and Bacillus subtilis. FEMS Microbiol Ecol 48:249–259
Rosario MM, Fredrick KL, Ordal GW, Helmann JD (1994) Chemotaxis in Bacillus subtilis requires either of two functionally redundant CheW homologs. J Bacteriol 176:2736–2739
Rosario MML, Kirby JR, Bochar DA, Ordal GW (1995) Chemotactic methylation and behavior in Bacillus subtilis: role of two unique proteins, CheC and CheD. Biochemistry 34:3823–3831
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Saulmon MM, Karatan E, Ordal GW (2004) Effect of loss of CheC and other adaptational proteins on chemotactic behaviour in Bacillus subtilis. Microbiology 150:581–589
Shen M, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524
Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucl Acids Res 33:W244–W248
Tastan Bishop O, Kroon M (2011) Study of protein complexes via homology modeling, applied to cysteine proteases and their protein inhibitors. J Mol Model. doi:10.1007/s00894-011-0990-y
Usher KC, De La Cruz AFA, Dahlquist FW, James Remington S, Swanson RV, Simon MI (1998) Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability. Protein Sci 7:403–412
Van de Broek A, Lambrecht M, Vanderleyden J (1998) Bacterial chemotactic motility is important for the initiation of wheat root colonization by Azospirillum brasilense. Microbiology 144:2599–2606
Wallner B, Elofsson A (2003) Can correct protein models be identified? Protein Sci 12:1073–1086
Yao J, Allen C (2006) Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J Bacteriol 188:3697–3708
Yao W, Shi L, Liang D (2007) Crystal structure of scaffolding protein CheW from Thermoanaerobacter tengcongensis. Biochem Biophys Res Commun 361:1027–1032
Zhang W, Phillips GN (2003) Structure of the oxygen sensor in Bacillus subtilis: signal transduction of chemotaxis by control of symmetry. Structure 11:1097–1110
Zimmer MA, Tiu J, Collins MA, Ordal GW (2000) Selective methylation changes on the Bacillus subtilis chemotaxis receptor McpB promote adaptation. J Biol Chem 275:24264–24272
Zimmer MA, Szurmant H, Saulmon MM, Collins MA, Bant JS, Ordal GW (2002) The role of heterologous receptors in McpB-mediated signalling in Bacillus subtilis chemotaxis. Mol Microbiol 45:555–568
Acknowledgements
AY thanks Rhodes University and National Research Foundation (NRF) of South Africa for financial support, OR's work was funded by the NRF grant 71261. We thank Prof. R. Borriss (Humboldt University, Berlin) for providing us with newly sequenced B. amyloliquefaciens genomes. We also thank Prof. N. Bishop and Dr. T. Bray for kindly proofreading the manuscript. AY and OTB started this work at the Bioinformatics and Computational Biology Unit, University of Pretoria; then continued at Rhodes University, Grahamstown.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
ESM 1
(PDF 175 kb)
Rights and permissions
About this article
Cite this article
Yssel, A., Reva, O. & Tastan Bishop, O. Comparative structural bioinformatics analysis of Bacillus amyloliquefaciens chemotaxis proteins within Bacillus subtilis group. Appl Microbiol Biotechnol 92, 997–1008 (2011). https://doi.org/10.1007/s00253-011-3582-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3582-y