Abstract
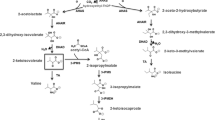
The Escherichia coli NADPH-dependent aldehyde reductase YqhD has contributed to a variety of metabolic engineering projects for production of biorenewable fuels and chemicals. As a scavenger of toxic aldehydes produced by lipid peroxidation, YqhD has reductase activity for a broad range of short-chain aldehydes, including butyraldehyde, glyceraldehyde, malondialdehyde, isobutyraldehyde, methylglyoxal, propanealdehyde, acrolein, furfural, glyoxal, 3-hydroxypropionaldehyde, glycolaldehyde, acetaldehyde, and acetol. This reductase activity has proven useful for the production of biorenewable fuels and chemicals, such as isobutanol and 1,3- and 1,2-propanediol; additional capability exists for production of 1-butanol, 1-propanol, and allyl alcohol. A drawback of this reductase activity is the diversion of valuable NADPH away from biosynthesis. This YqhD-mediated NADPH depletion provides sufficient burden to contribute to growth inhibition by furfural and 5-hydroxymethyl furfural, inhibitory contaminants of biomass hydrolysate. The structure of YqhD has been characterized, with identification of a Zn atom in the active site. Directed engineering efforts have improved utilization of 3-hydroxypropionaldehyde and NADPH. Most recently, two independent projects have demonstrated regulation of yqhD by YqhC, where YqhC appears to function as an aldehyde sensor.


Similar content being viewed by others
References
Atsumi S, Wu TY, Eckl EM, Hawkins SD, Buelter T, Liao JC (2010) Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl Microbiol Biotechnol 85(3):651–7
Babtie A, Tokuriki N, Hollfelder F (2010) What makes an enzyme promiscuous? Curr Opin Chem Biol 14(2):200–7
Berezina OV, Zakharova NV, Brandt A, Yarotsky SV, Schwarz WH, Zverlov VV (2010) Reconstructing the Clostridial n-butanol metabolic pathway in Lactobacillus brevis. Appl Microbiol Biotechnol 87(2):635–46
Claus P (1998) Selective hydrogenation of alpha, beta-unsaturated aldehydes and other C═O and C═C bonds containing compounds. Top Catal 5(1–4):51–62
Clomburg J, Gonzalez R (2010) Metabolic engineering of Escherichia coli for the production of 1, 2-propanediol from glycerol. Biotechnol Bioeng 103(1):148–161
Cornally D, Mee B, MacDonaill C, Tipton KF, Kelleher D, Windle HJ, Henehan GTM (2008) Aldo-keto reductase from Helicobacter pylori—role in adaptation to growth at acid pH. FEBS J 275(12):3041–50
Donaldson GK, Eliot AC, Flint D, Maggio-Hall LA, Nagarajan V (2007) Fermentative production of four carbon alcohols. WO 2007/050671 A2
Emptage M, Haynie SL, Laffend LA, Pucci JP, Whited G (2003) Process for the biological production of 1,3-propanediol with high titer. US 6514733 B1
Gallezot P, Richard D (1998) Selective hydrogenation of alpha, beta-unsaturated aldehydes. Catal Rev Sci Eng 40(1–2):81–126
Helbig K, Grosse C, Nies DH (2008) Cadmium toxicity in glutathione mutants of Escherichia coli. J Bacteriol 190(15):5439–54
Inui M, Suda M, Kimura S, Yasuda K, Suzuki H, Toda H, Yamamoto S, Okino S, Suzuki N, Yukawa H (2008) Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl Microbiol Biotechnol 77(6):1305–16
Jarboe LR, Hyduke DR, Tran LM, Chou KJY, Liao JC (2008) Determination of the Escherichia coli S-nitrosoglutathione response network using integrated biochemical and systems analysis. J Biol Chem 283(8):5148–57
Jarboe LR, Zhang XL, Wang X, Moore JC, Shanmugam KT, Ingram LO (2010) Metabolic engineering for production of biorenewable fuels and chemicals: contributions of synthetic biology. J Biomed Biotechnol. doi:10.1155/2010/761042
Johnson EA, Lin ECC (1987) Klebsiella pneumoniae 1, 3-propanediol-nad + oxidoreductase. J Bacteriol 169(5):2050–4
Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK (2007) Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl Environ Microbiol 73(13):4100–9
Lee C, Kim I, Lee J, Lee KL, Min B, Park C (2010) Transcriptional activation of the aldehyde reductase yqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12. J Bacteriol 192(16):4205–14
Lei J, Zhou YF, Li LF, Su XD (2009) Structural and biochemical analyses of yvgN and ytbE from Bacillus subtilis. Protein Sci 18(8):1792–800
Li HM, Chen J, Li YH (2008) Enhanced activity of YqhD oxidoreductase in synthesis of 1, 3-propanediol by error-prone PCR. Prog Nat Sci 18(12):1519–24
Liu Y, Tuysuz H, Jia CJ, Schwickardi M, Rinaldi R, Lu AH, Schmidt W, Schuth F (2010) From glycerol to allyl alcohol: iron oxide catalyzed dehydration and consecutive hydrogen transfer. Chem Commun 46(8):1238–40
Maness PC, Smolinski S, Blake DM, Huang Z, Wolfrum EJ, Jacoby WA (1999) Bactericidal activity of photocatalytic TiO2 reaction: toward an understanding of its killing mechanism. Appl Environ Microbiol 65(9):4094–8
Miller EN, Jarboe LR, Turner PC, Pharkya P, Yomano LP, York SW, Nunn D, Shanmugam KT, Ingram LO (2009a) Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl Environ Microbiol 75(19):6132–41
Miller EN, Jarboe LR, Yomano LP, York SW, Shanmugam KT, Ingram LO (2009b) Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl Environ Microbiol 75(13):4315–23
Miller EN, Turner PC, Jarboe LR, Ingram LO (2010) Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180. Biotechnol Lett 32(5):661–7
Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G (2004) Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci USA 101(3):745–50
Palosaari NR, Rogers P (1988) Purification and properties of the inducible coenzyme a-linked butyraldehyde dehydrogenase from Clostridium acetobutylicum. J Bacteriol 170(7):2971–6
Perez JM, Arenas FA, Pradenas GA, Sandoval JM, Vasquez CC (2008) Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J Biol Chem 283(12):7346–53
Rao Z, Ma Z, Shen W, Fang H, Zhuge J, Wang X (2008) Engineered Saccharomyces cerevisiae that produces 1, 3-propanediol from D-glucose. J Appl Microbiol 105(6):1768–76
Riehle MM, Bennett AF, Lenski RE, Long AD (2003) Evolutionary changes in heat-inducible gene expression in lines of Escherichia coli adapted to high temperature. Physiol Genomics 14(1):47–58
Riehle MM, Bennett AF, Long AD (2005) Changes in gene expression following high-temperature adaptation in experimentally evolved populations of E. coli. Physiol Biochem Zool 78(3):299–315
Rutherford BJ, Dahl RH, Price RE, Szmidt HL, Benke PI, Mukhopadhyay A, Keasling JD (2010) Functional genomics study of exogenous n-butanol stress in Escherichia coli. Appl Environ Microbiol 76(6):1935–45
Schutz H, Radler F (1984) Anaerobic reduction of glycerol to propanediol-1.3 by Lactobacillus brevis and Lactobacillus buchneri. Syst Appl Microbiol 5(2):169–78
Schwarzenbacher R, von Delft F, Canaves JM, Brinen LS, Dai XP, Deacon AM, Elsliger MA, Eshaghi S, Floyd R, Godzik A, Grittini C, Grzechnik SK, Guda C, Jaroszewski L, Karlak C, Klock HE, Koesema E, Kovarik JS, Kreusch A, Kuhn P, Lesley SA, McMullan D, McPhillips TM, Miller MA, Miller MD, Morse A, Moy K, Ouyang J, Page R, Robb A, Rodrigues K, Selby TL, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, Wang XH, West B, Wolf G, Hodgson KO, Wooley J, Wilson IA (2004) Crystal structure of an iron-containing 1,3-propanediol dehydrogenase (tm0920) from Thermotoga maritima at 1.3 angstrom resolution. Proteins Struct Funct Genet 54(1):174–7
Seo JW, Seo MY, Oh BR, Heo SY, Baek JO, Rairakhwada D, Luo LH, Hong WK, Kim CH (2010) Identification and utilization of a 1, 3-propanediol oxidoreductase isoenzyme for production of 1, 3-propanediol from glycerol in Klebsiella pneumoniae. Appl Microbiol Biotechnol 85(3):659–66
Shen CR, Liao JC (2008) Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab Eng 10(6):312–20
Soucaille P, Meynial-Salles I, Voelker F, Figge R (2008) Microorganisms and methods for production of 1,2-propanediol and acetol. WO 2008/116853 A1
Sulzenbacher G, Alvarez K, van den Heuvel RHH, Versluis C, Spinelli M, Campanacci V, Valencia C, Cambillau C, Eklund H, Tegoni M (2004) Crystal structure of E. coli alcohol dehydrogenase YqhD: Evidence of a covalently modified NADP coenzyme. J Mol Biol 342(2):489–502
Tang XM, Tan YS, Zhu H, Zhao K, Shen W (2009) Microbial conversion of glycerol to 1, 3-propanediol by an engineered strain of Escherichia coli. Appl Environ Microbiol 75(6):1628–34
Turner PC, Miller EN, Jarboe LR, Baggett CL, Shanmugam KT, Ingram LO (2010) YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance. J Ind Microbiol Biotech. doi:10.1007/s10295-010-0787-5
University of Oklahoma Gene Expression Database (2009) http://genexpdb.ou.edu/, accessed July 24th 2010
Villiers BRM, Hollfelder F (2009) Mapping the limits of substrate specificity of the adenylation domain of TycA. Chembiochem 10(4):671–82
Wang FH, Qu HJ, Zhang DW, Tian PF, Tan TW (2007) Production of 1, 3-propanediol from glycerol by recombinant E. coli using incompatible plasmids system. Mol Biotechnol 37:112–9
Zhu JG, Li S, Ji XJ, Huang H, Hu N (2009) Enhanced 1, 3-propanediol production in recombinant Klebsiella pneumoniae carrying the gene yqhD encoding 1, 3-propanediol oxidoreductase isoenzyme. World J Microbiol Biotechnol 25(7):1217–23
Acknowledgements
This work was funded by the Iowa State University Office of Biotechnology and the NSF Center for Biorenewable Chemicals Engineering Research Center, EEC-0813570. Chemical structures were obtained from ChemID Plus.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jarboe, L.R. YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl Microbiol Biotechnol 89, 249–257 (2011). https://doi.org/10.1007/s00253-010-2912-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2912-9