Abstract
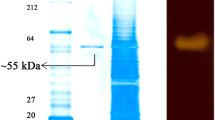
The increasing demand for novel biocatalysts stimulates exploration of resources from soil. Metagenomics, a culture independent approach, represent a sheer unlimited resource for discovery of novel biocatalysts from uncultured microorganisms. In this study, a soil-derived metagenomic library containing 90,700 recombinants was constructed and screened for lipase, cellulase, protease and amylase activity. A gene (pAMY) of 909 bp encoding for amylase was found after the screening of 35,000 Escherichia coli clones. Amino acid sequence comparison and phylogenetic analysis indicated that pAMY was closely related to uncultured bacteria. The molecular mass of pAMY was estimated about 38 kDa by sodium dodecyl sulphate polyacrylamide gel electrophoresis. Amylase activity was determined using soluble starch, amylose, glycogen and maltose as substrates. The maximal activity (2.46 U/mg) was observed at 40 °C under nearly neutral pH conditions with amylose; whereas it retains 90% of its activity at low temperature with all the substrates used in this study. The ability of pAMY to work at low temperature is unique for amylases reported so far from microbes of cultured and uncultured division.





Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aroonnual A, Takuya N, Si T, Watanalai P (2007) Role of several key residues in the catalytic activity of sucrose isomerase from Klebsiella pneumoniae NK33-98-8. Enzyme Microb Technol 40:1221–1227
Becks S, Bielawaski C, Henton D, Padala R, Burrows K, Slaby R (1995) Application of a liquid stable amylase reagent on the Ciba Corning express clinical chemistry system. Clin Chem 41:S186
Bertrand H, Poly F, Van VT, Lombard N, Nalin R, Vogel TM, Simonet P (2005) High molecular weight DNA recovery from soils prerequisite for biotechnological metagenomic library construction. J Microbiol Meth 62:1–11
Brady SF, Chao CJ, Handelsman J, Clardy J (2001) Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. OrgLett 3:1981–1984
Bordbar AK, Omidiyan K, Hosseinzadeh R (2005) Study on interaction of a-amylase from Bacillus subtilis with cetyl trimethylammonium bromide. Colloids Surf B: Biointerfaces 40:67–71
Choo DW, Kurihara T, Suzuki T, Soda K, Esaki N (1998) A coldadapted lipase of an Alaskan psychrotroph, Pseudomonas sp. strain B11-1: gene cloning and enzyme purification and characterization. Appl Environ Microbiol 64:486–491
Cottrell MT, Moore JA, Kirchman DL (1999) Chitinases from uncultured marine microorganisms. Appl Environ Microbiol 65:2552–2557
Elend C, Schmeisser C, Leggewie C, Babiak P, Carballeira JD, Steele HL, Reymond JL, Jaeger KE, Streit WR (2006) Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl Environ Microbiol 72:3637–3645
Gabor EM, Alkema WB, Janssen DB (2004) Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol 6:79–86
Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, Handelsman J (2002) Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl Environ Microbiol 68:4301–4306
Gupta R, Beg QK, Lorenz P (2002) Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol 59:15–32
Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B (2003) Microbial a-amylases: a biotechnological perspective. Process Biochem 38:1599–1616
Henne A, Daniel R, Schmitz RA, Gottschalk G (1999) Construction of environmental DNA Libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol 65:3901–3907
Henne A, Schmitz RA, Bömeke M, Gottschalk G, Daniel R (2000) Screening of environmental dna libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl Environ Microbiol 66:3113–3116
Johannes TW, Zhao H (2006) Directed evolution of enzymes and biosynthetic pathways. Curr Opin Microbiol 9:261–267
Lee MH, Lee CH, Oh TK, Song JK, Yoon JH (2006) Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl Environ Microbiol 72:7406–7409
Majerník A, Gottschalk G, Daniel R (2001) Screening of environmental DNA libraries for the presence of genes conferring Na+(Li+)/H +antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J Bacteriol 183:6645–6653
Miller GL (1959) Use of dinitrosalycylic acid for determination of reducing sugar. Anal Chem 31:426–428
Niemi RM, Heikkila MP, Lahti K, Kalso S, Niemela SI (2001) Extraction and purification of DNA in rhizosphere soil samples for PCR-DGGE analysis of bacterial consortia. J Microbiol Methods 45:155–165
Pandey A, Nigam P, Soccol CR, Soccol VT, Singh D, Mohan R (2000) Advances in microbial amylases. Appl Biochem 31:135–152
Ranjan R, Grover A, Kapardar RK, Sharma R (2005) Isolation of novel lipolytic genes from uncultured bacteria of pond water. Biochem Biophys Res Comm 335:57–65
Rhee JK, Ahn DG, Kim YG, Oh JW (2005) New thermophilic and thermostable esterase with sequence similarity to the hormonesensitive lipase family, cloned from a metagenomic library. Appl Environ Microbiol 71:817–825
Richardson TH, Tan X, Frey G, Callen W, Cabell M, Lam D, Macomber J, Short JM, Robertson DE, Miller C (2002) A novel, high performance enzyme for starch liquefaction discovery and optimization of a low pH, thermostable alpha amylase. J Biol Chem 277:26501–26507
Robertson DE, Chaplin JA, DeSantis G, Podar M, Madden M, Chi E, Richardson T, Milan A, Miller M, Weiner DP, Wong K, McQuaid J, Farwell B, Preston LA, Tan X, Snead MA, Keller M, Mathur E, Kretz PL, Burk MJ, Short JM (2004) Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol 70:2429–2436
Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, Macneil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66:2541–2547
Sandaa RA, Torsvik A, Enger O, Daae FL, Castberg T, Hahn D (1999) Analysis of bacterial communities in heavy metal-contaminated soils at different levels of resolution. FEMS Microbiol Ecol 30:237–251
Schmeisser C, Steele H, Streit WR (2007) Metagenomics,biotechnology with non-culturable microbes. Appl Microbiol Biotechnol 75:955–962
Sivaramakrishnan S, Gangadharan D, Nampoothiri KM, Soccol CR, Pandey A (2006) a-amylases from microbial sources—an overview on recent developments. Food Technol Biotechnol 44:173–184
Skerman VB (1969) Abstracts of microbiological methods. Wiley-Interscience, New York
Stach JEM, Bathe S, Clapp JP, Burns RG (2001) PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiology Ecol 36:139–151
Steele HL, Streit WR (2005) Metagenomics: advances in ecology and biotechnology. FEMS Microbiol Lett 247:105–111
Stoscheck CM (1990) Quantitation of protein. Methods Enzymol 182:50–69
Solbak AI, Richardson TH, McCann RT, Kline KA, Bartnek F, Tomlinson G, Tan X, Parra-Gessert L, Frey GJ, Podar M, Luginbühl P, Gray KA, Mathur EJ, Robertson DE, Burk MJ, Hazlewood GP, Short JM, Kerovuo J (2005) Discovery of pectin-degrading enzymes and directed evolution of a novel pectate lyase for processing cotton fabric. J Biol Chem 280:9431–9438
Voget S, Leggewie C, Uesbeck A, Raasch C, Jaeger KE, Streit WR (2003) Prospecting for novel biocatalysts in a soil metagenome. Appl Environ Microbiol 69:6235–6242
Yebra MJ, Blasco A, Sanz P (1999) Expression and secretion of Bacillus polymyxa neopullulanase in Saccharomyces cerevisiae. FEMS Microbiol Lett 170:41–49
Yun J, Kang S, Park S, Yoon H, Kim MJ, Heu S, Ryu S (2004) Characterization of a novel amylolytic enzyme encoded by a gene from a soil-derived metagenomic library. Appl Environ Microbiol 70:7229–7235
Acknowledgement
Sarika Sharma is thankful to Council of Scientific and Industrial Research (CSIR), Government of India for the award of Senior Research fellowship and Sumit Gandhi for help with sequence analysis and manuscript preparation. This work was financially supported by CSIR, New Delhi, project number NWP006.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharma, S., Khan, F.G. & Qazi, G.N. Molecular cloning and characterization of amylase from soil metagenomic library derived from Northwestern Himalayas. Appl Microbiol Biotechnol 86, 1821–1828 (2010). https://doi.org/10.1007/s00253-009-2404-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2404-y