Abstract
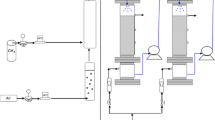
The performance and microbiology of two inorganic biofilters treating dimethyl sulphide (DMS) in the presence and absence of methanol was investigated. Addition of methanol was shown to result in an increase in DMS removal for methanol loadings below 90 g MeOH per cubic metre per hour with the optimal methanol loading around 10–15 g MeOH per cubic metre per hour for a DMS loading of 3.4 g DMS per cubic metre per hour, a fivefold increase in the DMS removal rate compared to the biofilter treating DMS alone. Microbial community analysis revealed that the addition of methanol led to a significant increase of up to an order of magnitude in the abundance of Hyphomicrobium spp. in the biofilter co-treating DMS and methanol compared to the biofilter treating DMS alone, whilst there was no significant difference in the abundance of Thiobacillus spp. between the two biofilters. Given the behaviour of the biofilter co-treating DMS and methanol, the magnitude of the increase in Hyphomicrobium spp. in the biofilter co-treating DMS and methanol and the ability of Hyphomicrobium spp. to use both methanol and DMS as growth substrates, it was concluded that Hyphomicrobium spp. were the microorganisms responsible for the bulk of the DMS degradation in the biofilter co-treating DMS and methanol.








Similar content being viewed by others
References
Andersson Chan A (2006) Attempted biofiltration of reduced sulphur compounds from a pulp and paper mill in northern Sweden. Environ Prog 25:152–160
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741
Budwill K, Coleman, RN (2002) Removal of dimethyl sulfide vapors in peat-based biofilters. Proceedings of the 95th AWMA Annual Meeting and Exhibition. Baltimore, MD
Cho KS, Hirai M, Shoda M (1991) Degradation characteristics of hydrogen sulfide, methanethiol, dimethyl sulfide, and dimethyl disulfide by Thiobacillus thioparus DW44 isolated from peat biofilter. J Ferment Bioeng 71:384–389
Cho KS, Hirai M, Shoda M (1992) Degradation of hydrogen sulfide by Xanthomonas sp. strain DY44 isolated from peat. Appl Environ Microbiol 58:1183–1189
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35:D169–D172
De Bont JAM, van Dijken JP, Harder W (1981) Dimethyl sulphoxide and dimethyl sulfide as a carbon, sulphur and energy source for growth of Hyphomicrobium S. J Gen Microbiol 127:315–323
Easter C, Quigley C, Burrowes P, Witherspoon J, Apgar D (2005) Odor and air emissions control using biotechnology for both collection and wastewater systems. Chem Eng Journ 113:93–104
Friedrich U, Prior K, Altendorf K, Lipski A (2002) High bacterial diversity of a waste gas-degrading community in an industrial biofilter as shown by a 16S rDNA clone library. Environ Microbiol 4:721–734
Friedrich U, Van Langenhove H, Altendorf K, Lipski A (2003) Microbial community and physicochemical analysis of an industrial waste gas biofilter and design of 16S rRNA-targeting oligonucleotide probes. Environ Microbiol 5:183–201
Harder W, Atwood MM (1978) Biology, physiology and biochemistry of hyphomicrobia. Adv Microbiol Physiol 17:303–359
Hartikainen T, Martikainen PJ, Olkkonen M, Ruuskanen J (2002) Peat biofilters in long-term experiments for removing odorous sulfur compounds. Water Air Soil Poll 133:335–348
Hirai M, Ohtake M, Shoda M (1990) Removal kinetics of hydrogen sulfide, methanethiol and dimethyl sulfide by peat biofilters. J Ferment Bioeng 70:334–339
Ho K-L, Chung Y-C, Lin Y-H, Tseng C-P (2007) Microbial populations analysis and field application of biofilter for the removal of volatile-sulfur compounds from swine wastewater treatment system. J Haz Mat 152:580–588
Kanagawa T, Kelly DP (1986) Breakdown of dimethyl sulphide by mixed cultures and by Thiobacillus thioparus. FEMS Microbiol Let 34:13–19
Layton AC, Karanth PN, Lajoie CA, Meyers AJ, Gregory IR, Stapleton D, Taylor DE, Sayler GS (2000) Quantification of Hyphomicrobium populations in activated sludge from and industrial wastewater treatment system as determined by 16S rRNA analysis. Appl Environ Microbiol 66:1167–1174
Luo J, Agnew MP (2001) Gas characteristics before and after biofiltration treating odorous emissions from animal rendering processes. Environ Technol 22:1091–1103
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nadkarni MA, Martin FE, Jacques NA, Hunter N (2002) Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiol 148:257–266
Okabe S, Satoh H, Watanabe Y (1999) In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol 65:3182–3191
Okabe S, Naitoh H, Satoh H, Watanabe Y (2002) Structure and function of nitrifying biofilms as determined by molecular techniques and the use of microelectrodes. Water Sci Technol 46:233–241
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci 12:357–358
Park SJ, Cho KS, Hirai M, Shoda M (1993) Removability of malodorous gases from a night soil treatment plant by a pilot-scale peat biofilter inoculated with Thiobacillus thioparus DW44. J Ferment Bioeng 76:55–59
Paungfoo C, Prasertsan P, Burrell PC, Intrasungkha N, Blackall LL (2006) Nitrifying bacterial communities in an aquaculture wastewater treatment system using fluorescence in situ hybridization (FISH), 16S rRNA gene cloning and phylogenetic analysis. Biotechnol Bioeng 97:985–990
Pol A, den Camp O, Huub JM, Mees SGM, Kersten MASH, van der Drift C (1994) Isolation of a dimethylsulfide-utilizing Hyphomicrobium species and its application in biofiltration of polluted air. Biodegradation 5:105–112
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sakano Y, Kerkhof L (1998) Assessment of changes in microbial community structure during operation of an ammonia biofilter with molecular tools. Appl Environ Microbiol 64:4877–4882
Sercu B, Núñex D, Van Langenhove H, Aroca G, Verstraete W (2005) Operational and microbiological aspects of a two-stage biotrickling filter removing hydrogen sulfide and dimethyl sulfide. Biotechnol Bioeng 90:259–269
Sercu B, Boon N, Vander Beken S, Verstraete W, Van Langenhove H (2006) Performance and microbial analysis of defined and non-defined inocula for the removal of dimethyl sulfide in a biotrickling filter. Biotechnol Bioeng 96:661–672
Shoda M (1991) Removal of dimethyl disulfide by the peat seeded with night soil sludge. J Ferment Bioeng 71:289–291
Smet E, Van Langenhove H, Verstraete W (1996) The effect of inoculation and the type of carrier material used on the biofiltration of methyl sulfides. Appl Microbiol Biotechnol 45:293–298
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuc Aci Res 25:4876–4882
Visscher PT, Taylor BF (1993) A new mechanism for the aerobic catabolism of dimethyl sulfide. Appl Environ Microbiol 59:3784–3789
Vitolins MI, Swaby RJ (1969) Activity of sulphur-oxidizing microorganisms in some Australian soils. Aust J Soil Res 7:171–183
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Zhang L, Hirai M, Shoda M (1991a) Removal characteristics of dimethyl sulfide, methanethiol and hydrogen sulfide by Hyphomicrobium sp. I55 isolated from peat biofilter. J Ferment Bioeng 73:392–396
Zhang L, Kuniyoshi I, Hirai M, Shoda M (1991b) Oxidation of dimethyl sulfide by Pseudomonas acidovorans DMR-11 isolated from peat biofilter. Biotech Let 13:223–228
Zhang L, Hirai M, Shoda M (1992) Removal characteristics of dimethyl sulfide, methanethiol and hydrogen sulfide by Hyphomicrobium sp. I55 and Pseudomonas acidovorans DMR-11. J Ferment Bioeng 74:174–178
Zhang Y, Liss SN, Allen DG (2006) The effects of methanol on the biofiltration of dimethyl sulfide in inorganic biofilters. Biotechnol Bioeng 95:734–743
Zhang Y, Liss SN, Allen DG (2007a) Effect of methanol on pH and stability of inorganic biofilters treating dimethyl sulfide. Environ Sci Technol 41:3752–3757
Zhang Y, Liss SN, Allen DG (2007b) Enhancing and modeling the biofiltration of dimethyl sulfide under dynamic methanol addition. Chem Eng Sci 62:2474–2481
Zhang Y, Liss SN, Allen DG (2008) Modeling the biofiltration of dimethyl sulfide in the presence of methanol in inorganic biofilters at steady state. Biotechnol Prog 24:845–851
Acknowledgements
The financial support of the Natural Sciences and Engineering Research Council of Canada (NSERC) and the research consortium “Minimizing the Impact of Pulp and Paper Mill Discharges” consisting of Aracruz Celulose S.A., Carter Holt Harvey Pulp and Paper, Domtar Inc., Eka Chemicals Inc., Georgia-Pacific Corporation, Irving Pulp and Paper Ltd., Japan Carlit Co. Ltd., ERCO Worldwide and Tembec Inc. is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hayes, A.C., Zhang, Y., Liss, S.N. et al. Linking performance to microbiology in biofilters treating dimethyl sulphide in the presence and absence of methanol. Appl Microbiol Biotechnol 85, 1151–1166 (2010). https://doi.org/10.1007/s00253-009-2272-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2272-5