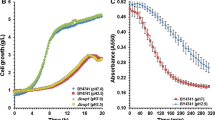
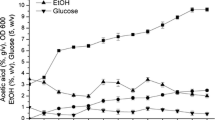
Abstract
The responses and adaptation mechanisms of the industrial Saccharomyces cerevisiae to vacuum fermentation were explored using proteomic approach. After qualitative and quantitative analyses, a total of 106 spots corresponding to 68 different proteins were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The differentially expressed proteins were involved in amino acid and carbohydrate metabolisms, various signal pathways (Ras/MAPK, Ras–cyclic adenosine monophosphate, and HOG pathway), and heat shock and oxidative responses. Among them, alternations in levels of 17 proteins associated with carbohydrate metabolisms, in particular, the upregulations of proteins involved in glycolysis, trehalose biosynthesis, and the pentose phosphate pathway, suggested vacuum-induced redistribution of the metabolic fluxes. The upregulation of 17 heat stress and oxidative response proteins indicated that multifactors contributed to oxidative stresses by affecting cell redox homeostasis. Taken together with upregulation in 14-3-3 proteins levels, 22 proteins were detected in multispots, respectively, indicating that vacuum might have promoted posttranslational modifications of some proteins in S. cerevisiae. Further investigation revealed that the elevations of the differentially expressed proteins were mainly derived from vacuum stress rather than the absence of oxygen. These findings provide new molecular mechanisms for understanding of adaptation and tolerance of yeast to vacuum fermentation.










Similar content being viewed by others
References
Atala DIP, Maugeri F (2005) Experimental development, instrumentation and control of a novel vacuum extractive fermentation process for ethanol production. J Biotechnol 118:S106–S106
Blomberg A, Adler L (1992) Physiology of osmotolerance in fungi. Adv Microb Physiol 33:145–212
Boucherie H, Dujardin G, Kermorgant M, Monribot C, Slonimski P, Perrot M (1995) Two-dimensional protein map of Saccharomyces cerevisiae: construction of a gene-protein index. Yeast 11:601–613
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brown SW, Oliver SG, Harrison DEF, Righelato RC (1981) Ethanol inhibition of yeast growth and fermentation: differences in the magnitude and complexity of the effect. Appl Microbiol Biotechnol 11:151–155
Bruckmann A, Hensbergen PJ, Balog CI, Deelder AM, de Steensma HY, van Heusden GP (2007) Post-transcriptional control of the Saccharomyces cerevisiae proteome by 14-3-3 proteins. J Proteome Res 6:1689–1699
Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92:351–366
Caesar R, Blomberg A (2004) The stress-induced Tfs1p requires NatB-mediated acetylation to inhibit carboxypeptidase Y and to regulate the protein kinase A pathway. J Biol Chem 279:38532–38543
Cheng JS, Yuan YJ (2006) Proteomic analysis reveals the spatial heterogeneity of immobilized Taxus cuspidata cells in support matrices. Proteomics 6:2199–2207
Cheng JS, Qiao B, Yuan YJ (2008) Comparative proteome analysis of robust Saccharomyces cerevisiae insights into industrial continuous and batch fermentation. Appl Microbiol Biotechnol 81:327–338
Cysewski GR, Wilke CR (1977) Rapid ethanol fermentations using vacuum and cell recycle. Biotechnol Bioeng 19:1125–1143
de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455:1251–1254
de Groot MJ, Daran-Lapujade P, van Breukelen B, Knijnenburg TA, de Hulster EA, Reinders MJ, Pronk JT, Heck AJ, Slijper M (2007) Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology 153:3864–3878
Ding MZ, Cheng JS, Xiao WH, Yuan YJ (2009) Comparative metabolomic analysis on industrial continuous and batch ethanol fermentation processes by GC-TOF-MS. Metabolomics 5:229–238
Garay-Arroyo A, Covarrubias AA, Clark I, Nino I, Gossett G, Martinez A (2004) Response to different environmental stress conditions of industrial and laboratory Saccharomyces cerevisiae strains. Appl Microbiol Biotechnol 63:734–741
Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391
Godon C, Lagniel G, Lee J, Buhler JM, Kieffer S, Perrot M, Boucherie H, Toledano MB, Labarre J (1998) The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem 273:22480–22489
Graves T, Narendranath NV, Dawson K, Power R (2007) Interaction effects of lactic acid and acetic acid at different temperatures on ethanol production by Saccharomyces cerevisiae in corn mash. Appl Microbiol Biotechnol 73:1190–1196
Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics 4:310–327
Hansen R, Pearson SY, Brosnan JM, Meaden PG, Jamieson DJ (2006) Proteomic analysis of a distilling strain of Saccharomyces cerevisiae during industrial grain fermentation. Appl Microbiol Biotechnol 72:116–125
Heux S, Cadiere A, Dequin S (2008) Glucose utilization of strains lacking PGI1 and expressing a transhydrogenase suggests differences in the pentose phosphate capacity among Saccharomyces cerevisiae strains. FEMS Yeast Res 8:217–224
Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66:300–372
Hu Y, Wang G, Chen GY, Fu X, Yao SQ (2003) Proteome analysis of Saccharomyces cerevisiae under metal stress by two-dimensional differential gel electrophoresis. Electrophoresis 24:1458–1470
Izawa S, Maeda K, Miki T, Mano J, Inoue Y, Kimura A (1998) Importance of glucose-6-phosphate dehydrogenase in the adaptive response to hydrogen peroxide in Saccharomyces cerevisiae. Biochem J 330:811–817
Jouhten P, Rintala E, Huuskonen A, Tamminen A, Toivari M, Wiebe M, Ruohonen L, Penttilä M, Maaheimo H (2008) Oxygen dependence of metabolic fluxes and energy generation of Saccharomyces cerevisiae CEN.PK113-1A. BMC Sys Bio 2:60
Kakiuchi K, Yamauchi Y, Taoka M, Iwago M, Fujita T, Ito T, Song SY, Sakai A, Isobe T, Ichimura T (2007) Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry 46:7781–7792
Kolkman A, Olsthoorn MM, Heeremans CE, Heck AJ, Slijper M (2005) Comparative proteome analysis of Saccharomyces cerevisiae grown in chemostat cultures limited for glucose or ethanol. Mol Cell Proteomics 4:1–11
Kolkman A, Daran-Lapujade P, Fullaondo A, Olsthoorn MM, Pronk JT, Slijper M, Heck AJ (2006) Proteome analysis of yeast response to various nutrient limitations. Mol Syst Biol 2:1–16
Lahti R, Kolakowski LF Jr, Heinonen J, Vihinen M, Pohjanoksa K, Cooperman BS (1990) Conservation of functional residues between yeast and E. coli inorganic pyrophosphatases. Biochim Biophys Acta 1038:338–345
Lloyd D, Morrell S, Carlsen HN, Degn H, James PE, Rowlands CC (1993) Effects of growth with ethanol on fermentation and membrane fluidity of Saccharomyces cerevisiae. Yeast 9:825–833
Maiorella BL, Blanch HW, Wilke CR (1983) By-product inhibition effects on ethanolic fermentation by Saccharomyces cerevisiae. Biotechnol Bioeng 25:103–121
Maiorella BL, Blanch HW, Wilke CR (1984) Economic evaluation of alternative ethanol fermentation processes. Biotechnol Bioeng 26:1003–1025
Nie Y, Liu H, Du G, Chen J (2008) Acetate yield increased by gas circulation and fed-batch fermentation in a novel syntrophic acetogenesis and homoacetogenesis coupling system. Bioresour Technol 99:2989–2995
Pahlman AK, Granath K, Ansell R, Hohmann S, Adler L (2001) The yeast glycerol 3-phosphatases Gpp 1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J Biol Chem 276:3555–3563
Pham TK, Wright PC (2008a) Proteomic analysis of calcium alginate-immobilized Saccharomyces cerevisiae under high-gravity fermentation conditions. J Proteome Res 7:515–525
Pham TK, Wright PC (2008b) The proteomic response of Saccharomyces cerevisiae in very high glucose conditions with amino acid supplementation. J Proteome Res 7:4766–4774
Pham TK, Chong PK, Gan CS, Wright PC (2006) Proteomic analysis of Saccharomyces cerevisiae under high gravity fermentation conditions. J Proteome Res 5:3411–3419
Pinto Mariano A, Bastos Borba Costa C, de Franceschi de Angelis D, Maugeri Filho F, Pires Atala DI, Wolf Maciel MR, Maciel Filho R (2008) Optimization strategies based on sequential quadratic programming applied for a fermentation process for butanol production. Appl Biochem Biotechnol. doi:https://doi.org/10.1007/s12010-008-8450-6
Pronk JT, Yde Steensma H, Van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607–1633
Querol A, Fernández-Espinar MT, del Olmo M, Barrio E (2003) Adaptive evolution of wine yeast. Int J Food Microbiol 86:3–10
Ragu S, Faye G, Iraqui I, Masurel-Heneman A, Kolodner RD, Huang ME (2007) Oxygen metabolism and reactive oxygen species cause chromosomal rearrangements and cell death. Proc Natl Acad Sci USA 104:9747–9752
Ramalingham A, Finn RK (1977) The vacuferm process: a new approach to fermentation alcohol. Biotechnol Bioeng 19:583–589
Rand JD, Grant CM (2006) The thioredoxin system protects ribosomes against stress-induced aggregation. Mol Biol Cell 17:387–401
Rep M, Proft M, Remize F, Tamás M, Serrano R, Thevelein JM, Hohmann S (2001) The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol 40:1067–1083
Roberts RL, Mösch HU, Fink GR (1997) 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 89:1055–1065
Rodrigues-Pousada CA, Nevitt T, Menezes R, Azevedo D, Pereira J, Amaral C (2004) Yeast activator proteins and stress response: an overview. FEBS Lett 567:80–85
Santivarangkna C, Kulozik U, Foerst P (2006) Effect of carbohydrates on the survival of Lactobacillus helveticus during vacuum drying. Lett Appl Microbiol 42:271–276
Santos PM, Simões T, Sá-Correia I (2009) Insights into yeast adaptive response to the agricultural fungicide mancozeb: a toxicoproteomics approach. Proteomics 9:657–670
Skoneczna A, Micialkiewicz A, Skoneczny M (2007) Saccharomyces cerevisiae Hsp31p, a stress response protein conferring protection against reactive oxygen species. Free Radic Biol Med 42:1409–1420
Stirling PC, Srayko M, Takhar KS, Pozniakovsky A, Hyman AA, Leroux MR (2007) Functional interaction between phosducin-like protein 2 and cytosolic chaperonin is essential for cytoskeletal protein function and cell cycle progression. Mol Biol Cell 18:2336–2345
Taherzadeh MJ, Niklasson C, Liden G (1997) Acetic acid-friend or foe in anaerobic batch conversion of glucose to ethanol by Saccharomyces cerevisiae? Chem Eng Sci 52:2653–2659
Trabalzini L, Paffetti A, Scaloni A, Talamo F, Ferro E, Coratza G, Bovalini L, Lusini P, Martelli P, Santucci A (2003) Proteomic response to physiological fermentation stresses in a wild-type wine strain of Saccharomyces cerevisiae. Biochem J 370(Pt 1):35–46
Ursic D, Sedbrook JC, Himmel KL, Culbertson MR (1994) The essential yeast Tcp1 protein affects actin and microtubules. Mol Biol Cell 5:1065–1080
Visser W, van der Baan AA, Batenburg-van der Vegte W, Scheffers WA, Krämer R, van Dijken JP (1994) Involvement of mitochondria in the assimilatory metabolism of anaerobic Saccharomyces cerevisiae cultures. Microbiology 140:3039–3046
Wang W, Sun J, Hartlep M, Deckwer WD, Zeng AP (2003) Combined use of proteomic analysis and enzyme activity assays for metabolic pathway analysis of glycerol fermentation by Klebsiella pneumoniae. Biotechnol Bioeng 83:525–536
Wegele H, Haslbeck M, Reinstein J, Buchner J (2003) Sti1 is a novel activator of the Ssa proteins. J Biol Chem 278:25970–25976
Winderickx J, de Winde JH, Crauwels M, Hino A, Hohmann S, Van Dijck P, Thevelein JM (1996) Regulation of genes encoding subunits of the trehalose synthase complex in Saccharomyces cerevisiae: novel variations of STRE-mediated transcription control? Mol Gen Genet 252:470–482
Wiseman A (2005) Avoidance of oxidative-stress perturbation in yeast bioprocesses by proteomic and genomic biostrategies? Lett Appl Microbiol 40:37–43
Wu CY, Bird AJ, Winge DR, Eide DJ (2007) Regulation of the yeast TSA1 peroxiredoxin by ZAP1 is an adaptive response to the oxidative stress of zinc deficiency. J Biol Chem 282:2184–2195
Xia JM, Yuan YJ (2009) Comparative lipidomics of four strains of Saccharomyces cerevisiae reveals different responses to furfural, phenol, and acetic acid. J Agric Food Chem 57:99–108
Yin Z, Stead D, Selway L, Walker J, Riba-Garcia I, McLnerney T, Gaskell S, Oliver SG, Cash P, Brown AJ (2004) Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics 4:2425–2436
Yoda K, Kawada T, Kaibara C, Fujie A, Abe M, Hitoshi H, Shimizu J, Tomishige N, Noda Y, Yamasaki M (2000) Defect in cell wall integrity of the yeast Saccharomyces cerevisiae caused by a mutation of the GDP-mannose pyrophosphorylase gene VIG9. Biosci Biotechnol Biochem 64:1937–1941
Zelenaya-Troitskaya O, Perlman PS, Butow RA (1995) An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J 14:3268–3276
Zhang R, Ou HY, Zhang CT (2004) DEG: a database of essential genes. Nucleic Acids Res 32(Database issue):D271–D272
Acknowledgments
The authors are grateful for the financial support from the National Natural Science Foundation of China (Key Program Grant No. 20736006), the National Basic Research Program of China (“973” Program 2007CB714301), Key Projects in the National Science and Technology Pillar Program (No. 2007BAD42B02), and the National Natural Science Foundation of China (No. 20706044).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 2101 kb)
Rights and permissions
About this article
Cite this article
Cheng, JS., Zhou, X., Ding, MZ. et al. Proteomic insights into adaptive responses of Saccharomyces cerevisiae to the repeated vacuum fermentation. Appl Microbiol Biotechnol 83, 909–923 (2009). https://doi.org/10.1007/s00253-009-2037-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2037-1