Abstract
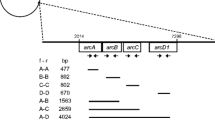
Oenococcus oeni is the most important lactic acid bacteria of the winemaking process involved in malolactic fermentation. Most O. oeni strains are able to catabolyze arginine via the arginine deiminase (ADI) pathway. The arcR, A, B, C, D1, and D2 cluster of O. oeni bacteria has been characterized. Here, we completed the ADI locus sequence. Downstream of arcD2 gene, we found an additional gene which encodes a putative arginyl-tRNA synthetase (argS2). It is not the same arginyl-tRNA synthetase which was sequenced in O. oeni MCW strain. Transcriptional analyses have shown that argS2 was induced by arginine. In addition, systematic polymerase chain reaction amplification of each arc gene and argS2 has provided a characteristic feature of the ADI locus within the O. oeni species: all genes of ADI locus are present or absent according to the strains.



Similar content being viewed by others
References
Barcelona-Andres B, Marina A, Rubio V (2002) Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J Bacteriol 184:6289–6300
Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A (2001) The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11:731–753
Champomier-Vergès MC, Zúñiga M, Morel-Deville F, Pérez-Martínez G, Zagorec M, Ehrlich SD (1999) Relationship between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol Lett 180:297–304
Cunin R, Glansdorff A, Piénard A, Stalon V (1986) Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50:314–352
Davey G, Heap H (1993) Appearence of the arginine phenotype in Lactococcus lactis subsp. cremoris 2204 following phage transduction. Can J Microbiol 39:154–158
D'Hooghe I, Vander Wauven C, Michiels J, Tricot C, de Wilde P, Vanderleyden J, Stalon V (1997) The arginine deiminase pathway in Rhizobium elti DNA sequence analysis and functional study of the arcABC genes. J Bacteriol 179:7403–7409
Dicks LM, Dellaglio F, Collins MD (1995) Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int J Syst Bacteriol 45:395–397
Divol B, Tonon T, Morichon S, Gindreau E, Lonvaud-Funel A (2003) Molecular characterization of Oenococcus oeni genes encoding proteins involved in arginine transport. J Appl Microbiol 94:738–746
Dong Y, Chen Y-YM, Snyder JA, Burne RA (2002) Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl Environ Microbiol 68:5549–5553
Gamper M, Zimmermann A, Hass D (1991) Anaerobic regulation of transcription initiation of the arcDABC operon of Pseudomonas aeruginosa. J Bacteriol 173:4742–4750
Gamper M, Ganter B, Polito MR, Haas D (1992) RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J Mol Biol 226:943–957
Kumar S, Tamura K, Nei M (1994) MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci 10:189–191
Lehtonen P (1996) Determination of amines and amino acids in wine. Am J Enol Vitic 47:127–133
Lonvaud-Funel A (1999) Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Van Leeuwenhoek 76:317–331
Lucas P, Landete J, Coton M, Coton E, Lonvaud-Funel A (2003) The tyrosine decarboxylase operon of Lactobacillus brevis IOEB 9809: characterization and conservation in tyramine-producing bacteria. FEMS Microbiol Lett 229:65–71
Maghnouj A, de Sousa Cabral TF, Stalon V, Vander Wauven C (1998) The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J Bacteriol 180:6468–6475
Maghnouj A, Abu-Bakr AAW, Baumberg S, Stalon V, Vander Wauven C (2000) Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol Lett 191:227–234
Martinez-Murcia AJ, Harland NM, Collins MD (1993) Phylogenetic analysis of some leuconostocs and related organisms as determined from large-subunit rRNA gene sequences: assessment of congruence of small- and large-subunit rRNA derived trees. J Appl Bacteriol 74:532–541
Martinis SA, Plateau P, Cavarelli J, Florentz C (1999) Aminoacyl-tRNA synthetases: a new image for a classical family. Biochimie 81:683–700
Mercenier A, Stalon V, Simon JP, Haas D (1982) Mapping of the arginine deiminase gene in Pseudomonas aeruginosa. J Bacteriol 149:787–788
Ohtani K, Bando M, Swe T, Banu S, Oe M, Hayashi H, Shimizu T (1997) Collagenase gene (colA) is located in the 3′-flanking region of the perfringolysin O (pfoA) locus in Clostridium perfringens. FEMS Microbiol Lett 146:155–159
Tonon T, Lonvaud-Funel A (2000) Metabolism of arginine and its positive effect on growth and revival of Oenococcus oeni. J Appl Microbiol 89:526–531
Tonon T, Bourdineaud JP, Lonvaud-Funel A (2001) The arcABC gene cluster encoding the arginine deiminase pathway of Oenococcus oeni, and arginine induction of a CRP-like gene. Res Microbiol 152:653–661
Zúñiga M, Champomier-Vergès M, Zagorec M, Pérez-Martínez G (1998) Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J Bacteriol 180:4154–4159
Zúñiga M, Miralles MC, Pérez-Martínez (2002) The product of arcR, the sixth gene of the arc operon of Lactobacillus sakei, is essential for expression of the arginine deiminase pathway. Appl Environ Microbiol 68:6051–6058
Acknowledgements
The authors are grateful to Dr. Caroline Duchaine for useful discussions. This work was supported by a grant from the Ministère de l'Education Nationale, de la Recherche et de la Technologie. M. Angelica Ganga was the recipient of a Connicyt-French Embassy (Chile) fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nehmé, B., Ganga, M.A. & Lonvaud-Funel, A. The arginine deiminase locus of Oenococcus oeni includes a putative arginyl-tRNA synthetase ArgS2 at its 3′-end. Appl Microbiol Biotechnol 70, 590–597 (2006). https://doi.org/10.1007/s00253-005-0095-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-0095-6