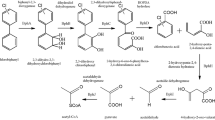
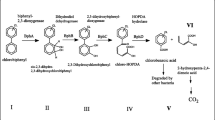
Abstract
Polychlorinated biphenyls (PCBs) are serious environmental pollutants that threaten both the natural ecosystem and human health. For remediation of environments contaminated with PCBs, several approaches that exploit the potential of microbes to degrade PCBs have been developed. These approaches include improvement of PCB solubilization and entry into the cell, pathway and enzyme engineering, and control of enzyme expression. In this mini-review, we briefly summarize these strategies and provide potentially useful knowledge for the further improvement of the bacterial breakdown of PCBs.




Similar content being viewed by others
References
Abraham WR, Nogales B, Golyshin PN, Pieper DH, Timmis KN (2002) Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr Opin Microbiol 5:246–253
Barriault D, Plante MM, Sylvestre M (2002) Family shuffling of a targeted bphA region to engineer biphenyl dioxygenase. J Bacteriol 184:3794–3800
Bartels F, Backhaus S, Moore ER, Timmis KN, Hofer B (1999) Occurrence and expression of glutathione-S-transferase-encoding bphK genes in Burkholderia sp. strain LB400 and other biphenyl-utilizing bacteria. Microbiology 145:2821–2834
Beltrametti F, Reniero D, Backhaus S, Hofer B (2001) Analysis of transcription of the bph locus of Burkholderia sp. strain LB400 and evidence that the ORF0 gene product acts as a regulator of the bphA1 promoter. Microbiology 147:2169–2182
Billingsley KA, Backus SM, Ward OP (1999) Effect of surfactant solubilization on biodegradation of polychlorinated biphenyl congeners by Pseudomonas LB400. Appl Microbiol Biotechnol 52:255–260
Bodour AA, Drees KP, Maier RM (2003) Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid Southwestern soils. Appl Environ Microbiol 69:3280–3287
Bruhlmann F, Chen W (1999a) Transformation of polychlorinated biphenyls by a novel BphA variant through the meta-cleavage pathway. FEMS Microbiol Lett 179:203–208
Bruhlmann F, Chen W (1999b) Tuning biphenyl dioxygenase for extended substrate specificity. Biotechnol Bioeng 63:544–551
Chen W, Bruhlmann F, Richins RD, Mulchandani A (1999) Engineering of improved microbes and enzymes for bioremediation. Curr Opin Biotechnol 10:137–141
Dai S, Vaillancourt FH, Maaroufi H, Drouin NM, Neau DB, Snieckus V, Bolin JT, Eltis LD (2002) Identification and analysis of a bottleneck in PCB biodegradation. Nat Struct Biol 9:934–939
Delawary M, Ohtsubo Y, Ohta A (2003) The dual functions of biphenyl-degrading ability of Pseudomonas sp. KKS102: energy acquisition and substrate detoxification. Biosci Biotechnol Biochem 67:1970–1975
Diaz E, Prieto MA (2000) Bacterial promoters triggering biodegradation of aromatic pollutants. Curr Opin Biotechnol 11:467–475
Erickson BD, Mondello FJ (1993) Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl Environ Microbiol 59:3858–3862
Evans BS, Dudley CA, Klasson KT (1996) Sequential anaerobic–aerobic biodegradation of PCBs in soil slurry microcosms. Appl Biochem Biotechnol 57/58:885–894
Fava F, Bertin L, Fedi S, Zannoni D (2003) Methyl-beta-cyclodextrin-enhanced solubilization and aerobic biodegradation of polychlorinated biphenyls in two aged contaminated soils. Biotechnol Bioeng 81:381–390
Furukawa K (1994) Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation 5:289–300
Furukawa K (2000) Engineering dioxygenases for efficient degradation of environmental pollutants. Curr Opin Biotechnol 11:244–249
Furukawa K (2003) ‘Super bugs’ for bioremediation. Trends Biotechnol 21:187–190
Gilbert ES, Crowley DE (1997) Plant compounds that induce polychlorinated biphenyl biodegradation by Arthrobacter sp. strain B1B. Appl Environ Microbiol 63:1933–1938
Gilmartin N, Ryan D, Sherlock O, Dowling D (2003) BphK shows dechlorination activity against 4-chlorobenzoate, an end product of bph-promoted degradation of PCBs. FEMS Microbiol Lett 222:251–255
Golyshin PM, Fredrickson HL, Giuliano L, Rothmel R, Timmis KN, Yakimov MM (1999) Effect of novel biosurfactants on biodegradation of polychlorinated biphenyls by pure and mixed bacterial cultures. New Microbiol 22:257–267
Grimm AC, Harwood CS (1997) Chemotaxis of Pseudomonas spp to the polyaromatic hydrocarbon naphthalene. Appl Environ Microbiol 63:4111–4115
Grimm AC, Harwood CS (1999) NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J Bacteriol 181:3310–3316
Harwood CS, Parales RE, Dispensa M (1990) Chemotaxis of Pseudomonas putida toward chlorinated benzoates. Appl Environ Microbiol 56:1501–1503
Hiraoka Y, Yamada T, Tone K, Futaesaku Y, Kimbara K (2002) Flow cytometry analysis of changes in the DNA content of the polychlorinated biphenyl degrader Comamonas testosteroni TK102: effect of metabolites on cell–cell separation. Appl Environ Microbiol 68:5104–5112
Hrywna Y, Tsoi TV, Maltseva OV, Quensen JF III, Tiedje JM (1999) Construction and characterization of two recombinant bacteria that grow on ortho- and para-substituted chlorobiphenyls. Appl Environ Microbiol 65:2163–2169
Keasling JD, Bang SW (1998) Recombinant DNA techniques for bioremediation and environmentally friendly synthesis. Curr Opin Biotechnol 9:135–140
Kim IS, Lee H, Trevors JT (2001) Effects of 2,2′,5,5′-tetrachlorobiphenyl and biphenyl on cell membranes of Ralstonia eutropha H850. FEMS Microbiol Lett 200:17–24
Kitagawa W, Suzuki A, Hoaki T, Masai E, Fukuda M (2001) Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1 demonstrated by denaturing gradient gel electrophoresis. Biosci Biotechnol Biochem 65:1907–1911
Kumamaru T, Suenaga H, Mitsuoka M, Watanabe T, Furukawa K (1998) Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat Biotechnol 16:663–666
Labbe D, Garnon J, Lau PC (1997) Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J Bacteriol 179:2772–2776
Lajoie CA, Layton AC, Sayler GS (1994) Cometabolic oxidation of polychlorinated biphenyls in soil with a surfactant-based field application vector. Appl Environ Microbiol 60:2826–2833
Law AM, Aitken MD (2003) Bacterial chemotaxis to naphthalene desorbing from a nonaqueous liquid. Appl Environ Microbiol 69:5968–5973
Makkar RS, Rockne KJ (2003) Comparison of synthetic surfactants and biosurfactants in enhancing biodegradation of polycyclic aromatic hydrocarbons. Environ Toxicol Chem 22:2280–2292
Master ER, Mohn WW (2001) Induction of bphA, encoding biphenyl dioxygenase, in two polychlorinated biphenyl-degrading bacteria, psychrotolerant Pseudomonas strain Cam-1 and mesophilic Burkholderia strain LB400. Appl Environ Microbiol 67:2669–2676
McKay DB, Seeger M, Zielinski M, Hofer B, Timmis KN (1997) Heterologous expression of biphenyl dioxygenase-encoding genes from a gram-positive broad-spectrum polychlorinated biphenyl degrader and characterization of chlorobiphenyl oxidation by the gene products. J Bacteriol 179:1924–1930
McKay DB, Prucha M, Reineke W, Timmis KN, Pieper DH (2003) Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus strain P6. J Bacteriol 185:2944–2951
Miyauchi K, Suh S-K, Nagata Y, Takagi M (1998) Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of γ-hexachlorocyclohexane by Sphingomonas paucimobilis. J Bacteriol 180:1354–1359
Mouz S, Merlin C, Springael D, Toussaint A (1999) A GntR-like negative regulator of the biphenyl degradation genes of the transposon Tn4371. Mol Gen Genet 262:790–799
Nandhagopal N, Yamada A, Hatta T, Masai E, Fukuda M, Mitsui Y, Senda T (2001) Crystal structure of 2-hydroxyl-6-oxo-6-phenylhexa-2,4-dienoic acid (HPDA) hydrolase (BphD enzyme) from the Rhodococcus sp. strain RHA1 of the PCB degradation pathway. J Mol Biol 309:1139–1151
Ohtsubo Y, Nagata Y, Kimbara K, Takagi M, Ohta A (2000) Expression of the bph genes involved in biphenyl/PCB degradation in Pseudomonas sp. KKS102 induced by the biphenyl degradation intermediate, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid. Gene 256:223–228
Ohtsubo Y, Delawary M, Kimbara K, Takagi M, Ohta A, Nagata Y (2001) BphS, a key transcriptional regulator of bph genes involved in polychlorinated biphenyl/biphenyl degradation in Pseudomonas sp. KKS102. J Biol Chem 276:36146–36154
Ohtsubo Y, Shimura M, Delawary M, Kimbara K, Takagi M, Kudo T, Ohta A, Nagata Y (2003) Novel approach to the improvement of biphenyl and polychlorinated biphenyl degradation activity: promoter implantation by homologous recombination. Appl Environ Microbiol 69:146–153
Pandey G, Jain RK (2002) Bacterial chemotaxis toward environmental pollutants: role in bioremediation. Appl Environ Microbiol 68:5789–5795
Pieper DH, Reineke W (2000) Engineering bacteria for bioremediation. Curr Opin Biotechnol 11:262–270
Ramos JL, Diaz E, Dowling D, de Lorenzo V, Molin S, O’Gara F, Ramos C, Timmis KN (1994) The behavior of bacteria designed for biodegradation. Biotechnology 12:1349–1356
Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A (2002) Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol 56:743–768
Reineke W (1998) Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu Rev Microbiol 52:287–331
Seah SY, Labbe G, Kaschabek SR, Reifenrath F, Reineke W, Eltis LD (2001) Comparative specificities of two evolutionarily divergent hydrolases involved in microbial degradation of polychlorinated biphenyls. J Bacteriol 183:1511–1516
Seeger M, Timmis KN, Hofer B (1995) Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for polychlorobiphenyl degradation encoded by the bph locus of Pseudomonas sp. strain LB400. Appl Environ Microbiol 61:2654–2658
Seeger M, Zielinski M, Timmis KN, Hofer B (1999) Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl Environ Microbiol 65:3614–3621
Shimizu S, Kobayashi H, Masai E, Fukuda M (2001) Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl Environ Microbiol 67:2021–2028
Shingler V (2003) Integrated regulation in response to aromatic compounds: from signal sensing to attractive behaviour. Environ Microbiol 5:1226–1241
Sikkema J, Bont JA de, Poolman B (1995) Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev 59:201–222
Singer AC, Gilbert ES, Luepromchai E, Crowley DE (2000) Bioremediation of polychlorinated biphenyl-contaminated soil using carvone and surfactant-grown bacteria. Appl Microbiol Biotechnol 54:838–843
Springael D, Kreps S, Mergeay M (1993) Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J Bacteriol 175:1674–1681
Suenaga H, Watanabe T, Sato M, Ngadiman, Furukawa K (2002) Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J Bacteriol 184:3682–3688
Takeda H, Yamada A, Miyauchi K, Masai E, Fukuda M (2004) Characterization of transcriptional regulatory genes for biphenyl degradation in Rhodococcus sp. strain RHA1. J Bacteriol 186:2134–2146
Tiedje JM, Quensen JF III, Chee-Sanford J, Schimel JP, Boyd SA (1993) Microbial reductive dechlorination of PCBs. Biodegradation 4:231–240
Timmis KN, Pieper DH (1999) Bacteria designed for bioremediation. Trends Biotechnol 17:200–204
Uragami Y, Senda T, Sugimoto K, Sato N, Nagarajan V, Masai E, Fukuda M, Mitsu Y (2001) Crystal structures of substrate free and complex forms of reactivated BphC, an extradiol type ring-cleavage dioxygenase. J Inorg Biochem 83:269–279
Vaillancourt FH, Han S, Fortin PD, Bolin JT, Eltis LD (1998) Molecular basis for the stabilization and inhibition of 2,3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol. J Biol Chem 273:34887–34895
Vaillancourt FH, Labbe G, Drouin NM, Fortin PD, Eltis LD (2002) The mechanism-based inactivation of 2,3-dihydroxybiphenyl 1,2-dioxygenase by catecholic substrates. J Biol Chem 277:2019–2027
Vicente M, Chater KF, De Lorenzo V (1999) Bacterial transcription factors involved in global regulation. Mol Microbiol 33:8–17
Watanabe T, Inoue R, Kimura N, Furukawa K (2000) Versatile transcription of biphenyl catabolic bph operon in Pseudomonas pseudoalcaligenes KF707. J Biol Chem 275:31016–31023
Watanabe T, Fujihara H, Furukawa K (2003) Characterization of the second LysR-type regulator in the biphenyl-catabolic gene cluster of Pseudomonas pseudoalcaligenes KF707. J Bacteriol 185:3575–3582
Xun L, Topp E, Orser CS (1992) Glutathione is the reducing agent for the reductive dehalogenation of tetrachloro-p-hydroquinone by extracts from a Flavobacterium sp. Biochem Biophys Res Commun 182:361–366
Acknowledgements
This work was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology and the Ministry of Agriculture, Forestry, and Fisheries (HC-04-2323-1), Japan. Part of this work was performed in the Eco Molecular Science Research Program of RIKEN. Y.O. was financially supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists. Y.O. was also a special postdoctoral researcher at RIKEN.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ohtsubo, Y., Kudo, T., Tsuda, M. et al. Strategies for bioremediation of polychlorinated biphenyls. Appl Microbiol Biotechnol 65, 250–258 (2004). https://doi.org/10.1007/s00253-004-1654-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-004-1654-y