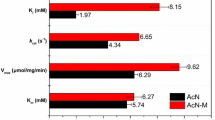
Abstract
The enzymatic hydrolysis of the nitrile group of different 2-acetoxynitriles was investigated in order to obtain catalysts that chemoselectively hydrolyse nitriles in the presence of ester groups. The biotransformation of four 2-acetoxynitriles [2-acetoxybutenenitrile (ABN), 2-acetoxyheptanenitrile (AHN), 2-acetoxy-2-(2-furyl)acetonitrile (AFN), and 2-acetoxy-2,3,3-trimethylbutanenitrile (ATMB)] by different bacterial strains that synthesise nitrilases or nitrile hydratases was studied. ABN, AHN and AFN were converted by various microorganisms belonging to different bacterial genera (e.g. Pseudomonas or Rhodococcus) expressing either nitrilase or nitrile hydratase activities. In contrast, no metabolism of the sterically hindered substrate ATMB was observed. All wild-type strains investigated formed considerable amounts of cyanide and aldehydes from the 2-acetoxynitriles. This indicated the presence of esterases converting the 2-acetoxynitriles to 2-hydroxynitriles, which then spontaneously decomposed to the corresponding aldehydes and cyanide. In order to suppress unwanted side-reactions, biotransformations were performed with recombinant Escherichia coli strains that heterologously expressed nitrilase activities originating from Pseudomonas, Rhodococcus, or Synechocystis strains. The attempted conversion of the 2-acetoxynitriles to almost stoichiometric amounts of the corresponding 2-acetoxycarboxylic acids was finally achieved by using either a recombinant E. coli strain that highly overexpressed the nitrilase gene from the pseudomonad or the purified enzyme derived from this strain.





Similar content being viewed by others
References
Bauer R, Hirrlinger B, Layh N, Stolz A, Knackmuss H-J (1994) Enantioselective hydrolysis of racemic 2-phenylpropionitrile and other (R,S)-2-arylpropionitriles by a new bacterial isolate, Agrobacterium tumefaciens strain d3. Appl Microbiol Biotechnol 42:1–7
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72:248–254
Bunch AW (1998) Nitriles. In: Rehm HJ, Reed G (eds) Biotechnology, vol 8a. Biotransformations I. Wiley-VCH Weinheim, pp 277–324
Effenberger F, Oßwald S (2001) Enantioselective hydrolysis of (RS)-2-fluoroarylacetonitriles using nitrilase from Arabidopsis thaliana. Tetrahedron Asymmetry 12:279–285
Effenberger F, Förster S, Wajant H (2000) Hydroxynitrile lyases in stereoselective catalysis. Curr Opin Biotechnol 11:532–539
Fukuda Y, Harada T, Izumi Y (1973) Formation of l-α-hydroxyacids from d,l-α-hydroxynitriles by Torulopsis candida GN405. J Ferment Technol 51:393–397
Gassman PG, Talley JJ (1978) Cyanohydrins—a general synthesis. Tetrahedron Lett 40:3773–3776
Goullet P (1973) An esterase zymogram of Escherichia coli. J Gen Microbiol 77:27–35
Goullet P, Picard B, Laget PF (1984) Purification and properties of carboxylesterase B of Escherichia coli. Ann Microbiol (Paris) 135A:375–387
Griengl H, Schwab H, Fechter M (2000) The synthesis of chiral cyanohydrins by oxynitrilases. Trends Biotechnol 18:252–256
Harper DB (1977) Microbial metabolism of aromatic nitriles. Enzymology of the C-N cleavage by Nocardia sp. (Rhodochrous group) N.C.I.B. 11216. Biochem J 165:309–319
Haruki M, Oohashi Y, Mizuguchi S, Matsuo Y, Morikawa M, Kanaya S. (1999) Identification of catalytically essential residues in Escherichia coli esterase by site-directed mutagenesis. FEBS Lett 454:262–266
Heinemann U, Engels D, Kiziak C, Mattes R, Stolz A (2003) Cloning, heterologous expression and enzymatic characterization of a nitrilase from the cyanobacterium Synechocystis spp. PCC6803. Appl Environ Microbiol 69: (in press)
Kakeya H, Sakai N, Sugai T, Ohta H (1991) Preparation of optically active α-hydroxy acid derivatives by microbial hydrolysis of cyanhydrins and its application to the synthesis of (R)-4-dodecanolide. Agric Biol Chem 55:1877–1881
Kiziak C (1998) Heterologe Expression der Nitrilase aus Pseudomonas fluorescens EBC191 und chimärer Enzymvarianten in E. coli. Reinigung, Stabilisierung und biochemische Charakterisierung der Enzyme. Diplomarbeit, Universität Stuttgart
Kobayashi M, Shimizu S (1994) Versatile nitrilases: nitrile-hydrolysing enzymes. FEMS Microbiol Lett 120:217–224
Kobayashi M, Yanaka N, Nagasawa T, Yamada H (1990) Purification and characterization of a novel nitrilase of Rhodococcus rhodochrous K22 that acts on aliphatic nitriles. J Bacteriol 172:4807–4815
Layh N, Stolz A, Förster S, Effenberger F, Knackmuss H-J (1992) Enantioselective hydrolysis of O-acetylmandelonitrile to O-acetylmandelic acid by bacterial nitrilases. Arch Microbiol 158:405–411
Layh N, Stolz A, Böhme J, Effenberger F, Knackmuss H-J (1994) Enantioselective hydrolysis of racemic naproxen nitrile and naproxen amide to S-naproxen by new bacterial isolates. J Biotechnol 33:175–182
Layh N, Hirrlinger B, Stolz A, Knackmuss H-J (1997) Enrichment strategies for nitriles hydrolysing bacteria. Appl Microbiol Biotechnol 47:668–674
Martínková L, Křen V (2002) Nitrile- and amide-converting microbial enzymes: stereo-, regio- and chemoselectivity. Biocatal Biotransform 20:79–93
Nagasawa T, Yamada H (1995) Microbial production of commodity chemicals. Pure Appl Chem 67:1241–1256
Nagasawa T, Mauger J, Yamada H (1990) A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3. Eur J Biochem 194:765–772
Peist R, Koch A, Bolek P, Sewitz S, Kolbus T, Boos W (1997) Characterization of the aes gene of Escherichia coli encoding an enzyme with esterase activity. J Bacteriol 179:7679–7686
Piotrowski M, Schönfelder S, Weiler EW (2001) The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode β-cyano-l-alanine hydratase/nitrilase. J Biol Chem 276:2616–2621
Raadt A de, Klempier N, Faber K, Griengl H (1992) Chemoselective enzymatic hydrolysis of aliphatic and alicyclic nitriles. J Chem Soc Perkin Trans I:137–140
Ress-Löschke M, Hauer B, Mattes R, Engels D (2000) Nitrilase aus Rhodococcus rhodochrous NCIMB 11216. German patent application No DE 10010149A1
Schulze B (2002) Hydrolysis and formation of C-N bonds. In: Drauz K, Waldmann H (eds) Enzyme catalysis in organic synthesis, vol II. Wiley-VCH, Weinheim, pp 699–715
Stevenson DE, Feng R, Dumas F, Groleau D, Mihoc A, Storer AC (1992) Mechanistic and structural studies on Rhodococcus ATCC 39484 nitrilase. Biotechnol Appl Biochem 15:283–302
Stumpp T, Wilms B, Altenbuchner J (2000) Ein neues, l-Rhamnose-induzierbares Expressionssystem für Escherichia coli. Biospektrum 6:33–36
Trott S, Bauer R, Knackmuss H-J, Stolz A (2001) Genetic and biochemical characterization of an enantioselective amidase from Agrobacterium tumefaciens d3. Microbiology 147:1815–1824
Yamamoto K, Fujimatsu I, Komatsu K-I (1992) Purification and characterization of the nitrilase from Alcaligenes faecalis ATCC 8750 responsible for enantioselective hydrolysis of mandelonitrile. J Ferment Bioeng 73:425–430
Zollner H (1989) Handbook of enzyme inhibitors. VCH, Weinheim, Germany
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Heinemann, U., Kiziak, C., Zibek, S. et al. Conversion of aliphatic 2-acetoxynitriles by nitrile-hydrolysing bacteria. Appl Microbiol Biotechnol 63, 274–281 (2003). https://doi.org/10.1007/s00253-003-1382-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-003-1382-8