Abstract
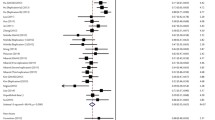
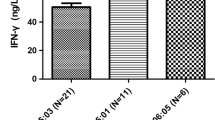
Hepatitis B virus (HBV) affects approximately 68 million people in China, and 10–15% of adults infected with HBV develop chronic hepatitis B, liver cirrhosis, liver failure or hepatocellular carcinoma (HCC). HLA-DPB1 gene polymorphism and expression have been shown to be associated with HBV infection susceptibility and spontaneous clearance. The aim of this study is to evaluate the role of HLA-DPB1 gene polymorphism in HBV infection. HLA-DPB1 and rs9277535 polymorphisms were investigated in 259 patients with HBV infection and 442 healthy controls (HCs) using sequence-based typing. The mRNA of HLA-DPB1 was measured by real-time polymerase chain reaction. HLA-DPB1 genes and rs9277535 polymorphisms were all associated with HBV infection in the Sichuan Han population. rs9277535A and HLA-DPB1*04:02 played a protective role against HBV infection. rs9277535G and DPB1*05:01 were associated with susceptibility to HBV infection. rs9277535GG had significantly higher HLA-DPB1 mRNA expression in the HBV infection group compared with the HC group. HLA-DPB1*05:01 and HLA-DPB1*21:01 had significantly lower mRNA expression in the HBV infection group compared with the HC group. The meta-analysis revealed that HLA-DPB1*02:01, HLA-DPB1*02:02, HAL-DPB1*04:01 and HLA-DPB1*04:02 protected against HBV infection, while HLA-DPB1*05:01, HLA-DPB1*09:01, and HLA-DPB1*13:01 were risk factors for susceptibility to HBV infection. HLA-DPB1*02:01, HLA-DPB1*02:02, and HLA-DPB1*04:01 were associated with HBV spontaneous clearance, while HLA-DPB1*05:01 was associated with chronic HBV infection. HLA-DPB1 alleles and rs9277535 have a major effect on the risk of HBV infection, and HBV infection is associated with lower HLA-DPB1 expression. HLA-DPB1 alleles have an important role in HBV susceptibility and spontaneous clearance.



Similar content being viewed by others
Data availability
All the data and materials supporting the conclusions were included in the main paper.
Abbreviations
- HBV:
-
Hepatitis B virus
- HLA:
-
Human leukocyte antigens
- APCs:
-
Antigen-presenting cells
- CHB:
-
Chronic HBV carriers
- HC:
-
Healthy controls
- HBsAg:
-
Hepatitis B surface antigen
- HBsAb:
-
Hepatitis B surface antibody
- HBeAg:
-
Hepatitis Be antigen
- HBeAb:
-
Hepatitis Be antibody
- HBcAb:
-
Hepatitis core antibody
- HCV:
-
Hepatitis C virus
- HIV:
-
Human immunodeficiency virus
- HWE:
-
Hardy–Weinberg equilibrium
References
Anczurowski M, Hirano N (2018) Mechanisms of HLA-DP antigen processing and presentation revisited. Trends Immunol 39:960–964
Cho SW, Cheong JY, Ju YS, Oh DH, Suh YJ, Lee KW (2008) Human leukocyte antigen class II association with spontaneous recovery from hepatitis B virus infection in Koreans: analysis at the haplotype level. J Korean Med Sci 23:838–844
Cui Y, Jia J (2013) Update on epidemiology of hepatitis B and C in China. J GastroenterolHepatol 28(Suppl 1):7–10
Doganay L, Fejzullahu A, Katrinli S, Yilmaz Enc F, Ozturk O, Colak Y, Ulasoglu C, Tuncer I, DinlerDoganay G (2014) Association of human leukocyte antigen DQB1 and DRB1 alleles with chronic hepatitis B. World J Gastroenterol 20:8179–8186
Donaldson PT, Ho S, Williams R, Johnson PJ (2001) HLA class II alleles in Chinese patients with hepatocellular carcinoma. Liver 21:143–148
Hiramatsu K, Matsuda H, Nemoto T, Nosaka T, Saito Y, Naito T, Takahashi K, Ofuji K, Ohtani M, Suto H et al (2017) Identification of novel variants in HLA class II region related to HLA DPB1 expression and disease progression in patients with chronic hepatitis C. J Med Virol
Hu L, Zhai X, Liu J, Chu M, Pan S, Jiang J, Zhang Y, Wang H, Chen J, Shen H, Hu Z (2012) Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology 55:1426–1431
Huang YH, Liao SF, Khor SS, Lin YJ, Chen HY, Chang YH, Huang YH, Lu SN, Lee HW, Ko WY et al (2020) Large-scale genome-wide association study identifies HLA class II variants associated with chronic HBV infection: a study from Taiwan Biobank. Aliment PharmacolTher 52:682–691
Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H et al (2009) A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 41:591–595
Katrinli S (2017) NilayKaratasErkut G, Ozdil K, Yilmaz Enc F, Ozturk O, Kahraman R, Tuncer I, DinlerDoganay G, Doganay L: HLA DPB1 15:01 allele predicts spontaneus hepatitis B surface antigen seroconversion. Acta Gastroenterol Belg 80:351–355
Li X, Zhou L, Gu L, Gu Y, Chen L, Lian Y, Huang Y (2017) Veritable antiviral capacity of natural killer cells in chronic HBV infection: an argument for an earlier anti-virus treatment. J Transl Med 15:220
Liao Y, Cai B, Li Y, Chen J, Tao C, Huang H, Wang L (2014) Association of HLA-DP/DQ and STAT4 polymorphisms with HBV infection outcomes and a mini meta-analysis. PLoS One 9:e111677
Mardian Y, Yano Y, Wasityastuti W, Ratnasari N, Liang Y, Putri WA, Triyono T, Hayashi Y (2017) Genetic polymorphisms of HLA-DP and isolated anti-HBc are important subsets of occult hepatitis B infection in Indonesian blood donors: a case-control study. Virol J 14:201
Matsuda H, Hiramatsu K, Akazawa Y, Nosaka T, Saito Y, Ozaki Y, Hayama R, Takahashi K, Naito T, Ofuji K et al (2018) Genetic polymorphism and decreased expression of HLA class II DP genes are associated with HBV reactivation in patients treated with immunomodulatory agents. J Med Virol 90:712–720
Niehrs A, Altfeld M (2020) Regulation of NK-cell function by HLA class II. Front Cell Infect Microbiol 10:55
Niehrs A, Garcia-Beltran WF, Norman PJ, Watson GM, Holzemer A, Chapel A, Richert L, Pommerening-Roser A, Korner C, Ozawa M et al (2019) A subset of HLA-DP molecules serve as ligands for the natural cytotoxicity receptor NKp44. Nat Immunol 20:1129–1137
Nishida N, Ohashi J, Khor SS, Sugiyama M, Tsuchiura T, Sawai H, Hino K, Honda M, Kaneko S, Yatsuhashi H et al (2016) Understanding of HLA-conferred susceptibility to chronic hepatitis B infection requires HLA genotyping-based association analysis. Sci Rep 6:24767
Nishida N, Ohashi J, Sugiyama M, Tsuchiura T, Yamamoto K, Hino K, Honda M, Kaneko S, Yatsuhashi H, Koike K et al (2015) Effects of HLA-DPB1 genotypes on chronic hepatitis B infection in Japanese individuals. Tissue Antigens 86:406–412
Nishida N, Sawai H, Kashiwase K, Minami M, Sugiyama M, Seto WK, Yuen MF, Posuwan N, Poovorawan Y, Ahn SH et al (2014) New susceptibility and resistance HLA-DP alleles to HBV-related diseases identified by a trans-ethnic association study in Asia. PLoS One 9:e86449
Nishida N, Sugiyama M, Sawai H, Nishina S, Sakai A, Ohashi J, Khor SS, Kakisaka K, Tsuchiura T, Hino K et al (2018) Key HLA-DRB1-DQB1 haplotypes and role of the BTNL2 gene for response to a hepatitis B vaccine. Hepatology 68:848–858
O’Brien TR, Kohaar I, Pfeiffer RM, Maeder D, Yeager M, Schadt EE, Prokunina-Olsson L (2011) Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun 12:428–433
Ou G, Xu H, Yu H, Liu X, Yang L, Ji X, Wang J, Liu Z (2018) The roles of HLA-DQB1 gene polymorphisms in hepatitis B virus infection. J Transl Med 16:362
Ou G, Liu X, Yang L, Yu H, Ji X, Liu F, Xu H, Qian L, Wang J, Liu Z (2019) Relationship between HLA-DPA1 mRNA expression and susceptibility to hepatitis B. J Viral Hepat 26:155–161
Paximadis M, Mathebula TY, Gentle NL, Vardas E, Colvin M, Gray CM, Tiemessen CT, Puren A (2012) Human leukocyte antigen class I (A, B, C) and II (DRB1) diversity in the black and Caucasian South African population. Hum Immunol 73:80–92
Petersdorf EW, Malkki M (2015) O’HUigin C, Carrington M, Gooley T, Haagenson MD, Horowitz MM, Spellman SR, Wang T, Stevenson P: High HLA-DP expression and graft-versus-host disease. N Engl J Med 373:599–609
Schone B, Bergmann S, Lang K, Wagner I, Schmidt AH, Petersdorf EW, Lange V (2018) Predicting an HLA-DPB1 expression marker based on standard DPB1 genotyping: linkage analysis of over 32,000 samples. Hum Immunol 79:20–27
Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ (2015) Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386:1546–1555
Shin HR, Oh JK, Masuyer E, Curado MP, Bouvard V, Fang YY, Wiangnon S, Sripa B, Hong ST (2010) Epidemiology of cholangiocarcinoma: an update focusing on risk factors. Cancer Sci 101:579–585
Thomas R, Thio CL, Apps R, Qi Y, Gao X, Marti D, Stein JL, Soderberg KA, Moody MA, Goedert JJ et al (2012) A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol 86:6979–6985
Wang FS, Fan JG, Zhang Z, Gao B, Wang HY (2014) The global burden of liver disease: the major impact of China. Hepatology 60:2099–2108
Wang LY, Chen CF, Wu TW, Lai SK, Chu CC, Lin HH (2019) Response to hepatitis B vaccination is co-determined by HLA-DPA1 and -DPB1. Vaccine 37:6435–6440
Yamashita Y, Anczurowski M, Nakatsugawa M, Tanaka M, Kagoya Y, Sinha A, Chamoto K, Ochi T, Guo T, Saso K et al (2017) HLA-DP(84Gly) constitutively presents endogenous peptides generated by the class I antigen processing pathway. Nat Commun 8:15244
Yan ZH, Fan Y, Wang XH, Mao Q, Deng GH, Wang YM (2012) Relationship between HLA-DR gene polymorphisms and outcomes of hepatitis B viral infections: a meta-analysis. World J Gastroenterol 18:3119–3128
Yu L, Cheng YJ, Cheng ML, Yao YM, Zhang Q, Zhao XK, Liu HJ, Hu YX, Mu M, Wang B et al (2015) Quantitative assessment of common genetic variations in HLA-DP with hepatitis B virus infection, clearance and hepatocellular carcinoma development. Sci Rep 5:14933
Zhang Q, Yin J, Zhang Y, Deng Y, Ji X, Du Y, Pu R, Han Y, Zhao J, Han X et al (2013) HLA-DP polymorphisms affect the outcomes of chronic hepatitis B virus infections, possibly through interacting with viral mutations. J Virol 87:12176–12186
Zhu M, Dai J, Wang C, Wang Y, Qin N, Ma H, Song C, Zhai X, Yang Y, Liu J et al (2016) Fine mapping the MHC region identified four independent variants modifying susceptibility to chronic hepatitis B in Han Chinese. Hum Mol Genet 25:1225–1232
Acknowledgments
The authors thank the participants for generously providing the venous blood samples.
Funding
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) under contract 2016-I2M-3–024, Funding of Sichuan Science and Technology Department under contract 2017RZ0047 and 2018SZ0395 and Ministry of Science and Technology of China, Grant/Award number: 2014EG150133. The CIFMS program provided us blood samples and relevant data, funding of Sichuan Science and Technology Department project provide us the cytokine detect methods and methods of HLA-DPB1 typing, program of Ministry of Science and Technology of China provide us HBV relevant detect methods.
Author information
Authors and Affiliations
Contributions
Ou GJ, Xu HX, and Ji X performed the experiments. Liu X collected and evaluated the samples. Ou GJ and Liu XJ wrote the original draft of the manuscript. Wang J and Liu XJ designed the experiments and performed the data analysis, discussed the results, and substantially revised the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethic committees of the Institution of Blood Transfusion, CAMS&PUMC, and was conducted according to the principles of the Declaration of Helsinki. All participants provided written informed consent before enrolment, and the study’s protocol was approved by the ethic committees of the Institution of Blood Transfusion, CAMS&PUMC.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ou, G., Liu, X., Xu, H. et al. Variation and expression of HLA-DPB1 gene in HBV infection. Immunogenetics 73, 253–261 (2021). https://doi.org/10.1007/s00251-021-01213-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-021-01213-w