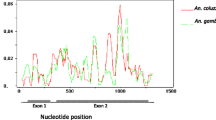
Abstract
Genes involved in host–pathogen interactions are often strongly affected by positive natural selection. The Duffy antigen, coded by the Duffy antigen receptor for chemokines (DARC) gene, serves as a receptor for Plasmodium vivax in humans and for Plasmodium knowlesi in some nonhuman primates. In the majority of sub-Saharan Africans, a nucleic acid variant in GATA-1 of the gene promoter is responsible for the nonexpression of the Duffy antigen on red blood cells and consequently resistance to invasion by P. vivax. The Duffy antigen also acts as a receptor for chemokines and is expressed in red blood cells and many other tissues of the body. Because of this dual role, we sequenced a ~3,000-bp region encompassing the entire DARC gene as well as part of its 5′ and 3′ flanking regions in a phylogenetic sample of primates and used statistical methods to evaluate the nature of selection pressures acting on the gene during its evolution. We analyzed both coding and regulatory regions of the DARC gene. The regulatory analysis showed accelerated rates of substitution at several sites near known motifs. Our tests of positive selection in the coding region using maximum likelihood by branch sites and maximum likelihood by codon sites did not yield statistically significant evidence for the action of positive selection. However, the maximum likelihood test in which the gene was subdivided into different structural regions showed that the known binding region for P. vivax/P. knowlesi is under very different selective pressures than the remainder of the gene. In fact, most of the gene appears to be under strong purifying selection, but this is not evident in the binding region. We suggest that the binding region is under the influence of two opposing selective pressures, positive selection possibly exerted by the parasite and purifying selection exerted by chemokines.




Similar content being viewed by others
References
Allison AC (2009) Genetic control of resistance to human malaria. Curr Opin Immunol 21:499–505
Baum J, Thomas AW, Conway DJ (2003) Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax. Genetics 163(4):1327–1336
Butcher GA (1996) Models for malaria: nature knows best. Parasitol Today 12(10):378–382
Butcher GA, Mitchell GH, Cohen S (2010) Plasmodium knowlesi infections in a small number of non-immune natural hosts (Macaca fascicularis) and in rhesus monkeys (M. mulatta). Trans R Soc Trop Med Hyg 104:75–77
Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton
Chaudhuri A, Polyakova J, Zbrzezna V, Pogo AO (1995) The coding sequence of Duffy blood group gene in humans and simians: restriction fragment length polymorphism, antibody and malarial parasite specificities, and expression in nonerythroid tissues in Duffy-negative individuals. Blood 85:615–621
Chitnis CE, Chaudhuri A, Horuk R, Pogo OA, Miller LH (1996) The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med 184:1531–1536
de Brevern AG, Wong H, Tournamille C, Colin Y, Le Van KC, Etchebest C (2005) A structural model of a seven-transmembrane helix receptor: the Duffy antigen/receptor for chemokine (DARC). Biochim Biophys Acta 1724(3):288–306
Demogines A, Truong KA, Sawyer SL (2012) Species-specific features of DARC, the primate receptor for Plasmodium vivax and Plasmodium knowlesi. Mol Biol Evol 29(2):445–449
Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, Lal AA (2005) A monkey’s tale: the origin of Plasmodium vivax as a human malaria parasite. Proc Natl Acad Sci U S A 102:1980–1985
Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genetics, University of Washington, Seattle. Distributed by the author
Fooden J (1994) Malaria in macaques. Int J Primatol 15(4):573–596
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hadley TJ, Peiper SC (1997) From malaria to chemokine receptor: the emerging physiologic role of the Duffy blood group antigen. Blood 89:3077–3091
Hamblin MT, Di Rienzo A (2000) Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet 66:1669–1679
Hamblin MT, Thompson EE, Di Rienzo A (2002) Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet 70:369–383
Harris EE, Meyer D (2006) The molecular signature of selection underlying human adaptations. Am J Phys Anthropol Suppl 43:89–130
Iwamoto S, Li J, Omi T, Ikemoto S, Kajii E (1996) Identification of a novel exon and spliced form of Duffy mRNA that is the predominant transcript in both erythroid and postcapillary venule endothelium. Blood 87:378–385
Iyer J, Gruner AC, Renia L, Snounou G, Preiser PR (2007) Invasion of host cells by malaria parasites: a tale of two protein families. Mol Microbiol 65:231–249
Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6:29
Langhi DM Jr, Bordin JO (2006) Duffy blood group and malaria. Hematology 11:389–398
Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK (1975) Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189:561–563
Miller LH, Mason SJ, Clyde DF, McGinniss MH (1976) The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med 295:302–304
Palatnik M, Rowe AW (1984) Duffy and Duffy-related human antigens in primates. J Hum Evol 13:173–179
Poirriez J, Dei-Cas E, Landau I (1994) Further description of blood stages of Plasmodium petersi from Cercocebus albigena monkey. Folia Parasitol 41(3):168–172
Prugnolle F, Ollomo B, Durand P, Yalcindag E, Arnathau C, Elguero E, Berry A, Pourrut X, Gonzalez JP, Nkoghe D, Akiana J, Verrier D, Leroy E, Ayala FJ, Renaud F (2011) African monkeys are infected by Plasmodium falciparum nonhuman primate-specific strains. Proc Natl Acad Sci U S A 108(29):11948–11953
Prugnolle F, Durand P, Neel C, Ollomo B, Ayala FJ, Arnathau C, Etienne L, Mpoudi-Ngole E, Nkoghe D, Leroy E, Delaporte E, Peeters M, Renaud F (2010) African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci U S A 107(4):1458–1463
Rayner JC, Liu W, Peeters M, Sharp PM, Hahn BH (2011) A plethora of Plasmodium species in wild apes: a source of human infection? Trends Parasitol 27(5):222–229
Sasseville VG, Diters RW (2008) Impact of infections and normal flora in nonhuman primates on drug development. ILAR J 49(2):179–190
Sawai H, Otani H, Arisue N, Palacpac N, de Oliveira ML, Pathirana S, Handunnetti S, Kawai S, Kishino H, Horii T, Tanabe K (2010) Lineage-specific positive selection at the merozoite surface protein 1 (msp1) locus of Plasmodium vivax and related simian malaria parasites. BMC Evol Biol 10:52
Schmidt LH, Fradkin R, Harrison J, Rossan RN (1977) Differences in the virulence of Plasmodium knowlesi for Macaca irus (fascicularis) of Philippine and Malayan origins. AmJTrop Med Hyg 26:612–622
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tournamille C, Blancher A, Le Van KC, Gane P, Apoil PA, Nakamoto W, Cartron JP, Colin Y (2004) Sequence, evolution and ligand binding properties of mammalian Duffy antigen/receptor for chemokines. Immunogenetics 55:682–694
Tournamille C, Le Van KC, Gane P, Blanchard D, Proudfoot AE, Cartron JP, Colin Y (1997) Close association of the first and fourth extracellular domains of the Duffy antigen/receptor for chemokines by a disulfide bond is required for ligand binding. J Biol Chem 272:16274–16280
Tung J, Primus A, Bouley AJ, Severson TF, Alberts SC, Wray GA (2009) Evolution of a malaria resistance gene in wild primates. Nature 460(7253):388–391
Wong WS, Yang Z, Goldman N, Nielsen R (2004) Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168:1041–1051
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Yang Z, Nielsen R (2002) Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19:908–917
Yang Z, Swanson WJ (2002) Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol Biol Evol 19(1):49–57
Acknowledgments
We thank the two anonymous reviewers for their helpful comments that helped us improve the article. This research was supported by a PSC CUNY 38 grant to EEH and a C3IRG award from City University of New York to EEH and CKJ as well as grants from INCTC/CNPq, CTC/CEPID/FAPESP, Regional Blood Center of Ribeirão Preto, and Millennium Institute/CNPq to WA Silva-Jr. The authors would like to thank Adriana Marques, Cristiane A. Pereira, and Anemare R. Dinarte for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 600 kb)
Rights and permissions
About this article
Cite this article
Oliveira, T.Y.K., Harris, E.E., Meyer, D. et al. Molecular evolution of a malaria resistance gene (DARC) in primates. Immunogenetics 64, 497–505 (2012). https://doi.org/10.1007/s00251-012-0608-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-012-0608-2