Abstract
The natural killer cell gene complex (NKC) encodes receptors belonging to the C-type lectin superfamily expressed primarily by NK cells and other leukocytes. In the rat, the chromosomal region that starts with the Nkrp1a locus and ends with the Ly49i8 locus is predicted to contain 67 group V C-type lectin superfamily genes, making it one of the largest congregation of paralogous genes in vertebrates. Based on physical proximity and phylogenetic relationships between these genes, the rat NKC can be divided into four major parts. We have previously reported the cDNA cloning of the majority of the genes belonging to the centromeric Nkrp1/Clr cluster and the two telomeric groups, the Klre1–Klri2 and the Ly49 clusters. Here, we close the gap between the Nkrp1/Clr and the Klre1–Klri2 clusters by presenting the cDNA cloning and transcription patterns of eight genes spanning from Cd69 to Dectin1, including the novel Clec2m gene. The definition, organization, and evolution of the rat NKC are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 1991, Yokoyama and co-workers showed that genes encoding the C-type lectin-like (CLEC) receptors NKR-P1 and Ly49 were genetically closely linked in the mouse and postulated the existence of a genetic region encoding functionally related C-type lectin superfamily (CLSF) members. As these receptors primarily were expressed by natural killer (NK) cells, it was named the NK cell gene complex (NKC) (Yokoyama et al. 1991). In the pursuit of identifying structural gene(s) associated with regulation of NK cell mediated alloreactivity in the rat, we subsequently identified the equivalent rat gene complex (Dissen et al. 1996). Remarkably, both in the mouse and the rat, the genetic distance between the Nkrp1a and the Ly49 genes was estimated to ∼0.5 cM, based on analyses of recombinant inbred strains (Yokoyama et al. 1991) and parental to F1 backcrosses (Dissen et al. 1996), respectively. Assuming average crossing-over frequencies, this would correspond to a physical distance of less than 1 Mb. However, the first physical map of the NKC, based on pulsed field gel electrophoresis, indicated the region to be much larger (Dissen et al. 1996). In the rat, other CLSF genes were rapidly mapped to the intervening region (Berg et al. 1998a, b; Dissen et al. 1997), followed by still more genes in the mouse and, with the identification of a human NKC, in the human (Renedo et al. 1997; Suto et al. 1997; Hamann et al. 1997; Colonna et al. 2000; Sobanov et al. 2001; Boles et al. 1999; Plougastel et al. 2001). The releases of the sequences amassed by the international genome projects finally exposed the real size of this gene complex in different species. Thus, aided by the Rat Genome project, we could report that the genetic region spanning from the most centromeric Nkrp1 to the most telomeric Ly49 gene covered 3.3 Mb, with a predicted content of 67 CLSF genes (Nylenna et al. 2005a). However, although the conservation of sequence features among the many intraspecific paralogs and interspecific orthologs has greatly facilitated prediction of novel CLSF genes, in silico prediction is error prone and uninformative as to whether the predicted genes are expressed.
Functional traits such as resistance to cytomegalovirus (reviewed in Webb et al. 2002) and fungal infection (reviewed in Sun and Zhao 2007) and association with celiac disease have been mapped to discrete genes in the NKC (Hue et al. 2004), and association to experimentally induced arthritis was mapped to the neighboring gene complex, APLEC, also encoding CLSF receptors (Flornes et al. 2004; Lorentzen et al. 2007). For other quantitative trait loci mapped to this chromosomal region, including loci controlling experimentally induced inflammatory responses in the rat, the associated structural genes await identification. An accurate and complete inventory of the genetic content of this genetic region would represent a useful tool in their ultimate identification, and thus of importance for studying pathogenetic mechanisms behind the human inflammatory and autoimmune diseases. We have previously described the cDNA cloning of the genes constituting the Klre1–Klri2 cluster (Berg et al. 1998a, b; Dissen et al. 1997; Saether et al. 2005; Westgaard et al. 2003) and, barring pseudogenes, most of the genes belonging to the Ly49 gene cluster (Naper et al. 2002; Nylenna et al. 2005b). Together, the two clusters make up the telomeric part of the rat NKC. From the centromeric end of the complex, we recently reported the cDNA cloning of the Nkrp1 genes and most of the Clr genes (Kveberg et al. 2009), leaving a gap between the most distal Clr gene and the Lox1 (Olr1) gene, previously reported by others (Nagase et al. 1998) and situated immediately centromeric to the Klre1–Klri2 cluster. Here, we close this gap by presenting the cDNA cloning and transcription patterns of eight genes spanning from Cd69 to Dectin1, including a novel gene which we have called Clec2m. We also include transcription analyses of the Nkrp1 and Clr genes and present the genomic organization of the 67 predicted group V CLSF localized in the rat NKC.
Materials and methods
cDNA cloning
As the genes reported here were cDNA-cloned before the availability of the rat genome sequence assembly, they were identified as described in Flornes et al. (2004) by (1) searching the GenBank rat Trace archive and the EST database for sequences homologous to the published mouse and human receptors, using the NCBI BLAST program and (2) performing pairwise BLAST on recently released partially or fully sequenced rat BAC clones. Gene-specific (nested) primers in the 5′- and 3′-untranslated regions (UTRs) were generated from the predicted sequences (primers shown in Supplemental Table 1). The 3′-UTR of Clec12a could not be identified, and this gene was therefore cDNA-cloned using RACE with nested 3′ primers from the GeneRacer™ kit (Invitrogen.com).
mRNA from mouse and rat lymph node, bone marrow, or testis were isolated with Dynabeads mRNA Direct kit (Invitrogen), and first-strand cDNA synthesis was carried out with M-MLV reverse transcriptase RNase H− (Promega) using 3–5 μl of the eluate in a 20-μl reaction volume. PCR was performed on first-strand cDNA using gene-specific primers and PfuTurbo DNA polymerase (Stratagene), and the products were cloned into pCR2.1-TOPO vector (Invitrogen). For every gene, three or more independent clones were sequenced. The sequences were analyzed using software supplied by the Norwegian EMBNet node at the Biotechnology Centre in Oslo, Norway.
Quantitative PCR
A panel of RNA from purified cells and tissues from PVG rats was prepared previously (Dissen et al. 1997). Briefly, cells and tissues were extracted by lysis in isothiocyanate (Gibco BRL) followed by ultracentrifugation on a cesium chloride (Gibco BRL) gradient and phenol/chloroform extractions. Dendritic cells were isolated by spontaneous migration from split ear-halves floating on RPMI for 48 h, testis were dissected from rats at day 4 and weeks 7.5, and 15. RNA from dendritic cells and testis were isolated with RNeasy (Qiagen), and first-strand cDNA synthesis for all samples was performed with Superscript II reverse transcriptase (Invitrogen) and random hexamers using 1 µg total RNA in a 30-µl reaction. Amplification was performed on an Applied Biosystem 7900HT Fast Real Time PCR system (PerkinElmer) using the 5′nuclease assay and qPCR SuperMix with Rox (Invitrogen). Analyses were done with the SDS2.1 software (Applied Biosystems, PerkinElmer). Primers and probes were designed using PrimerExpress 3.0 (Applied Biosystems, PerkinElmer) and purchased from Eurofins MWG Operon (Ebersberg, Germany). Individual samples were run in triplicate, and the relative quantity of RNA for each target was normalized to RNA from the reference gene plasma membrane calcium ATPase 4 (PMCA4).
Bioinformatics
Sequence similarity searches were performed with BLAST programs, running on the NCBI or Ensembl websites. The phylograms were constructed with NJ plot based on alignments with the ClustalX program, with the pileup program in the GCG package (Accelrys Inc., San Diego, CA, USA) and Bayesian inference methods (MrBayes—http://mrbayes.csit.fsu.edu)
Results and discussion
Sequences and transcription patterns
Eight genes spanning the gap between the Nkrp1/Clr genes and the Lox1 locus were cDNA cloned. In Fig. 1, the predicted amino acid sequences are presented together with their mouse and human counterparts, with closest sequence similarity (Fig. 2) and conserved chromosomal positions and orientation between the three species as orthology criteria.
Amino acid sequence alignments of the CLSF receptors. r rat, m mouse, h human. Dashes mean identity with top sequence. Points indicate gaps. Ex1, Ex2, etc. on top of alignment show start site of corresponding exons. Predicted transmembrane regions (TMpred) are underlined. ITAMs and ITIMs are indicated. Potential N-glycosylation sites are boxed. Membrane external cysteines are highlighted in yellow. Numbers connected with black lines on top of the CD69 and Dectin1 sequences indicate SS-bonds as demonstrated for crystal structures of human CD69 and mouse Dectin1 (http://www.ncbi.nlm.nih.gov/ Genbank/). For the other genes, the corresponding cysteines are similarly numbered. X above cysteines in Clec1a indicates probable extra disulphide bridge, which in addition to the bridge between the cysteines labeled 2, links the first α helix with the last β strand of the lectin-like domain. GenBank accession numbers: rat CD69–GU357488, rat Clec2m–GU357487, mouse Clec2m–GU553093, rat Clec12a–GU357484, rat Clec12b–GU357483, rat Clec1b–GU357482, rat Clec9a–GU357486, rat Clec1a–GU357481, rat Dectin-1–GU357485. The rat strains from which the sequence was derived are shown in parentheses (PVG, DA, or both). It should be noted that the reported coding sequences were identical to the corresponding parts of the genomic rat sequence based on the BN strain. The single exception was rat Dectin1, with two single base substitutions in exon 6, one silent and one missense giving rise to an A225G substitution (identical for four independent cDNAs sequenced)
Dendrogram of the novel receptors (plus Lox1) and their human and mouse orthologs (based on amino acid sequences). Numbers indicate bootstrap values (1,000 reiterations) and Bayesian clade credibility values. Values shown only for branching points with bootstrap values >900 or credibility values >90
Cd69 and Clec2m
Whereas the majority of group V CLSF genes consist of six coding exons, Cd69 and Clec2m have only five (Fig. 1). Rat Cd69 shows relatively high transcription levels, as measured by RT-PCR, in dendritic cells derived from skin (sDC), peritoneal macrophages (pMΦ), and resting B and T cells as well as ConA activated lymphocytes (Fig. 3). The transcription in resting lymphocytes is noteworthy. Cd69 was first cloned in the human (Ziegler et al. 1993), where it originally was reported as expressed primarily on activated lymphocytes and on NK cells, neutrophils, and platelets. The CD69 receptor has consequently been widely used as an in vivo as a marker for leukocyte activation and inflammatory responses. It has been extensively studied in the human and the mouse, where in vitro studies originally indicated proinflammatory function. More recent in vivo results indicate that it may act as a regulatory molecule, modulating inflammatory responses (reviewed in Sancho et al. 2005) as well as playing a role in lymphocyte trafficking, by interacting with the S1P1 receptor required for efficient egress of lymphocytes from the thymus and lymph nodes (Shiow et al. 2006) and immature B cells from the bone marrow (Allende et al. 2010).
Gene transcription as measured by quantitative PCR. The transcriptions of genes were calculated as the ratio of copy number of target gene to the copy number of endogenous reference gene, PMCA4. Thresholds for intermediate and high transcription were set to 1/100 and 1 relative to PMCA4. Not detected no PCR product detected after 40 cycles, DC dendritic cells, PM peritoneal macrophages, RNK16 NK cell line, LAK IL2-stimulated NK cells. Similar results were obtained using the HPRT1 gene (hypoxanthine guanine phosphoribosyl transferase 1) as the endogenous reference gene
Clec2m has, to our knowledge, not previously been cDNA cloned. Hao et al. (2006) labeled the in silico predicted rat and mouse gene Clec15a and Clec15ap, respectively, with the mouse gene predicted to be a pseudogene. We have cDNA cloned full-length versions of the gene both in the rat and the mouse (Fig. 1), but were unable to identify it in the human. As the term Clec15a is an established synonym for Klrg1 (Mafa), we propose the name Clec2m, denoting close kinship to the Clr (Clec2d) genes and high transcription in macrophages (see below). The gene shows highest transcription levels in sDC and pMΦ, and among tissues bone marrow and testis (Fig. 3).
Clec12a (Micl) and Clec12b
Clec12a possesses a single immunoreceptor tyrosine-based inhibition motif (ITIM) sequence (IxYxxL) in the cytoplasmic tail. Rat Clec12b contains the ITIM-like sequence AxYxxL, whereas mouse and human Clec12b both have the consensus ITIM VxYxxL (Fig. 1). For Clec12a, the inhibitory function suggested by the ITIM has been demonstrated both in the human, where it was first identified as myeloid inhibitory C-type lectin-like receptor (Marshall et al. 2004) and later in the mouse (reviewed in Huysamen and Brown 2009). In the human and the mouse (Pyz et al. 2008), Clec12a is expressed predominantly by DC, macrophages, and granulocytes and down-regulated following stimulation with select toll-like receptor ligands. In the rat, the highest transcription levels of Clec12a were found in sDC and pMΦ, and among tissues in bone marrow and brain. Less is known about the closely related Clec12b (reviewed in Huysamen and Brown 2009). Its transcription profile in the rat is distinct from that of Clec12a, with moderate transcription levels in sDC and neutrophils, no detectable transcription in pMΦ, and with strong transcription in the testis (Fig. 3).
Clec1b, Clec9a, and Dectin1
These receptors share an atypical immunoreceptor tyrosine-based activation motif (ITAM) D/ExYxxL in their cytoplasmic tail (Fig. 1). In the human and the mouse, all three have been shown to recruit and mediate activation signals via spleen tyrosine kinase (Syk) (Rogers et al. 2005; Fuller et al. 2007; Huysamen et al. 2008). Clec1b, first identified in the human and named CLEC-2 by Colonna et al. (2000), is expressed on various cell types including megakaryocytes and platelets, where it triggers platelet activation and aggregation (Suzuki-Inoue et al. 2006). Various exogenous and one endogenous ligand, the sialoglycoprotein podoplanin, have been identified (Suzuki-Inoue et al. 2007). In the rat, Clec1b transcription levels were moderate in neutrophils, low in lymphocytes, undetectable in sDC and pMΦ and high in spleen and liver (Fig. 3). Clec9a has recently been identified both in the human and the mouse as an activating receptor (Huysamen et al. 2008; Caminschi et al. 2008), expressed primarily by a subset of monocytes and the rare CD141+ subset of DC (Huysamen et al. 2008). In the rat, it shows high transcription in the spleen, moderate to low transcription levels in several other tissues and cell types, but no detectable transcription in sDC or CD4+ T cells (Fig. 3). Whereas the physiological roles of Clec1b and Clec9a remain unknown, much insight has been gained about roles of dendritic cell-associated C-type lectin 1 (Dectin1). It was first identified in the mouse (Ariizumi et al. 2000) and has been shown both in the mouse and in the human to function as the main leukocyte β-glucan binding receptor, with a major role in antifungal immunity (reviewed in Sun and Zhao 2007; Brown 2006). In the mouse, Dectin1 is most strongly expressed on monocytes, macrophages, neutrophils, and microglia, weakly on a T cell subset, and in the human also on B cells, mast cells, and eosinophils (Brown 2006). In the rat, Dectin1 was transcribed in all tissues and cell types tested (except the NK cell line RNK16), with particularly strong transcription in sDC, pMΦ, bone marrow, and the thymus (Fig. 3).
Clec1a
Finally, although the human Clec1a receptor was first published 10 years ago, under the name CLEC-1 (Colonna et al. 2000), little is known about its functional properties and roles. In the human, the cytoplasmic tail contains one tyrosine residue close to the N-terminus. This is also present in the mouse and the rat sequences, which have an additional tyrosine residue forming the pattern YxxTx13YxxT (Fig. 1). Whether the tyrosines are subject to phosphorylation is not known. A notable sequence feature of Clec1 is the two cysteines immediately preceding the cysteines predicted to form the disulphide bond labeled 2 in Fig. 1. This is a characteristic shared with the Ly49 receptors, where the two extra cysteines form an additional SS-bond between the α1 chain and the last β chain of the lectin domain. In the rat, Clec1a is transcribed in all cell types and tissues tested, with the strongest transcription in ConA-stimulated blasts, spleen and kidney.
Comments on transcription patterns
The presence of ITIM motifs in Clec12a (and possibly Clec12b) and (atypical) ITAM motifs in Clec1b and Clec9a suggests inhibitory and activating functions, as shown for the human and mouse orthologs. As the occurrence of closely related receptors with opposite signaling properties is suggestive of “paired opposing receptors,” their widely different transcription profiles are noteworthy. In Fig. 3, we have also included transcription analyses of the Clr and the Nkrp1 genes, as this information has not previously been published in the rat and is therefore needed for complete comparison of the expression programs of the rat NKC genes. The pattern of strong NK cell transcription exhibited by NKR-P1A, -B, and -F receptors is the rule for the majority of the rat NKC receptors previously reported. In contrast, most of the genes reported here, as well as most of the Clr genes, show modest to low or no transcription in NK cells, demonstrating a genetic content beyond the original definition of this large congregation of genes as an NK cell gene complex.
Phylogenetic analyses
A striking feature of the eight receptors reported is the wide range of sequence divergences between the rat, mouse, and human orthologs (Fig. 2). The most extreme examples are Clec1a, which is highly conserved, and Clec12a, which is extensively changed between the three species (Figs. 1 and 2). For Clec1a, the rat and mouse protein percent identity is 95.5 and the rodent versus human identities 71.3/69.4 (human-rat/human-mouse). For Clec12a, the corresponding figures are 73.3 and 47.7/50.6. The dissimilarities between the three Clec12a sequences suggest positive selection, a notion further strengthened by Ka/Ks analyses of the cDNA sequences. Ka/Ks analysis compares the rate of non-synonymous to synonymous substitutions, with a higher value indicating possible positive selection. For Clec1a, the mouse versus rat Ka/Ks ratio is 0.10 and for Clec12a, 1.00. Splitting the Clec12a sequences into exons 1–3, encoding the cytoplasmic tail/transmembrane/neck domains, and exons 3–6, encoding the lectin domain, i.e., the presumed ligand binding part of the receptor, gives Ka/Ks values of 0.57 and 1.42, respectively. The ligands of these receptors are unknown, but the analysis suggests that the Clec1a ligand is phylogenetically conserved, whereas the Clec12a receptors may be chasing a more rapidly evolving ligand.
Phylogenetic analyses indicate that CD69 and Clec2m belong to a different subfamily than the other six receptors (Fig. 2). On inclusion of the other CLSF receptors encoded by the rat NKC, they exhibit the closest sequence similarity with the Clr subfamily (Fig. 4). Although sensitive to gap parameters and not significant according to bootstrap analyses, the association seems reasonable considering the physical localization of Cd69 and Clec2m next to the Clr genes (Fig. 5) combined with the fact that they consist of only five coding exons, lacking a separate exon encoding the external membrane-proximal stalk. This is a property they share with the Clr genes, and contrasts with all the other NKC CLSF genes, which generally consist of six coding exons (apart from Lox1, which has eight coding exons).
Dendrogram based on deduced protein sequences of all the CLSF genes encoded by the rat NKC (only eight of the predicted 34 Ly49 receptors shown). Also included are Mafa1/Klrg1 and Mdl1/Clec5a, plus the seven predicted functional APLEC genes. Vertical black lines indicate major branches of the NKC receptors. The genes containing five coding exons are indicated with 5 ex. Only bootstrap values >900 from 1,000 reiterations are shown
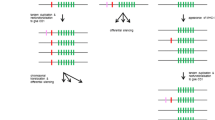
Map of the rat NKC. Arrows indicate gene orientation. Numbers below the line indicate distance from the starting point (the Nkrp1a gene at 165.637 Mb according to the current release of the rat genome). The four major clusters are color-coded: green Nkrp1/Clr gene cluster (including Cd69 and Clec2m), magenta Clec12a–Lox1 cluster, red Klre1–Klri2 cluster, blue Ly49 cluster. The topography of the chromosomal map with respect to major clusters is seen to be congruent with the major branches of the phylogenetic tree in Fig. 4. It should be emphasized that most of the genes shown here have been cDNA cloned primarily in the DA or PVG strain, so that the map shown here represents the positions of the predicted alleles from the BN strain, from which the rat genome sequence is derived. The extent of intestrain variation, including copy number variation, is so far largely uncharted
As for the other six genes, the Clec12 and Clec1 genes tended to cluster together (Fig. 2). The shape of the tree is, however, highly sensitive to gap parameters and to inclusion of other NKC genes (Fig. 4). The tree shape in Fig. 2 is similar to that reported by Hao et al. except for Clec9a, which was assigned to an entirely different clade of CLSF genes (Hao et al. 2006). Arguments in favor of including Clec9a in the Clec12 – Lox1 clade are the sharing of chromosomal localization and sequence features such as the atypical ITAM motif. Furthermore, a conserved feature of the NKC-encoded CLSF receptors is the Cx10C loop of amino acids clamped by an SS-bridge between the flanking cysteines labeled 1 in Fig. 1. In front of these, Clec1a and –b, Clec12a and –b, and Clec9a have two additional cysteines, the first encoded by a codon near the end of exon 3 (Fig. 1) and restricted to these receptors, whereas the second, encoded by the third codon in exon 4 (Fig. 1) is also present in CD94, Klrk1, the Nkg2 and the Klri receptors. The atypical ITAM motif and the two extra cysteines are likely to represent derived (apomorphic) rather than ancestral (plesiomorphic) states, so that the assigning of Clec9a to a different clade would implicate convergent evolution or sequence transfer through gene conversion. The simpler explanation is that their shared presence stems from phylogenetic kinship. Whether the extra cysteines are involved in inter- or intrachain disulphide bonding is not known.
The organization of genes in the rat NKC
The phylogenetic tree shown in Fig. 4 includes group II and group V CLSF members localized on rat chromosome 4 (for simplicity, only eight Ly49 genes are shown, as all 34 cluster together in a single clade). In addition to the genes depicted in Fig. 5, the tree includes Mafa1 (Klrg1), Mdl1 (Clec5a), and the seven genes grouped together in the APLEC cluster. The tree is seen to consist of six major branches. The exact branching is highly sensitive to gap parameters, but apart from the placement of Clec9a as discussed above, it is identical to the tree presented by Hao et al. (2006) with respect to major branches. The interesting feature of the tree is that the gene content of the major branches correlates exactly with the chromosomal clustering of the genes (apart from Mafa1 and Mdl1). In addition to supporting the soundness of the tree, the observation indicates that this large congregation of paralogous genes has evolved by local gene duplications without major reshufflings. At the local level chromosomal inversion events have occurred, as evidenced by the changes in gene orientations, as well as segmental duplications involving more than one gene in the case of the Ly49 gene cluster, as previously discussed (Nylenna et al. 2005b). A notable feature is the conservation of orientation of the 41 most telomeric genes of the complex, from Klrk1 to Ly49i8. The evolutionary events leading to the gene arrangement of the Nkrp1/Clr cluster seem particularly complex. In addition to shifting gene orientations, the cluster consist of two separate blocks containing Nkrp1 genes with opposite signaling function next to series of Clr genes with incongruent patterns of topographical versus sequence neighborness.
The demarcation of the NKC
The chromosomal region starting with Nkrp1a and ending with Ly49i8 contains almost all known group V CLSF genes in the rat. Exceptions are Cd72, localized on chromosome 5, and Klrg1 (Mafa1), Mdl1 (Clec5a), and Clec2l, all three on rat chromosome 4, at distances 7, 97, and 99 Mb centromeric to Nkrp1a, respectively. Furthermore, the region contains nothing but group V CLSF genes, with Gabarapl1 (Gamma-aminobutyric acid receptor-associated protein-like 1) as the single known exception. Although heterogeneous in gene content, it displays conserved gene organization and persistence of orthologous gene lineages across four major mammalian orders, as described by Hao et al. (2006). These authors also included in the NKC the Mincle–Dcir gene cluster (Fig. 5), which in all four orders occupy a chromosomal region close to, but clearly separate from the Nkrp1a–Ly49i8 region. When we first described the genetic region containing the Mincle–Dcir gene cluster, which we named the antigen-presenting cell lectin-like receptor gene complex (APLEC), we presented arguments for and against including these genes in the NKC. Counterarguments were (1) the evolutionary distance between the two groups of genes. Whereas the APLEC genes are classified as group II CLSF (http://www.imperial.ac.uk/research/animallectins/), with conserved amino acid residues involved in calcium-dependent saccharide binding (Weis et al. 1991), the group V CLSF have lost these amino acid motifs (Weis et al. 1998). (2) The distance separating the APLEC genes from the NKC. In the mouse and the rat, it is 5–6 Mb; in the human, cow, horse, and dog, only ∼1.5 Mb. In all these species, the intervening region is packed with non-CLSF genes, which would be included in the NKC if defined as encompassing the APLEC, with loss of communicative precision and a potential source of confusion in the future mapping of functional traits to this chromosomal region. Even when narrowly defined, the NKC represent one of the largest congregation of paralogous genes known in vertebrates.
Abbreviations
- APLEC:
-
Antigen-presenting cell lectin-like receptor gene complex
- CLEC:
-
C-type lectin-like
- CLSF:
-
C-type lectin superfamily
- sDC:
-
Skin dendritic cells
- ITAM:
-
Immunoreceptor tyrosine-based activation motif
- ITIM:
-
Immunoreceptor tyrosine-based inhibition motif
- NKC:
-
Natural killer cell gene complex
- pMΦ:
-
Peritoneal macrophages
- TM:
-
Transmembrane
References
Allende ML, Tuymetova G, Lee BG, Bonifacino E, Wu YP, Proia RL (2010) S1P1 receptor directs the release of immature B cells from bone marrow into blood. J Exp Med 207:1113–1124
Ariizumi K, Shen GL, Shikano S, Xu S, Ritter R III, Kumamoto T, Edelbaum D, Morita A, Bergstresser PR, Takashima A (2000) Identification of a novel, dendritic cell-associated molecule, dectin- 1, by subtractive cDNA cloning. J Biol Chem 275:20157–20167
Berg SF, Dissen E, Westgaard IH, Fossum S (1998a) Molecular characterization of rat NKR-P2, a lectin-like receptor expressed by NK cells and resting T cells. Int Immunol 10:379–385
Berg SF, Dissen E, Westgaard IH, Fossum S (1998b) Two genes in the rat homologous to human NKG2. Eur J Immunol 28:444–450
Boles KS, Barten R, Kumaresan PR, Trowsdale J, Mathew PA (1999) Cloning of a new lectin-like receptor expressed on human NK cells. Immunogenetics 50:1–7
Brown GD (2006) Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol 6:33–43
Caminschi I, Proietto AI, Ahmet F, Kitsoulis S, Shin TJ, Lo JC, Rizzitelli A, Wu L, Vremec D, van Dommelen SL, Campbell IK, Maraskovsky E, Braley H, Davey GM, Mottram P, van de Velde N, Jensen K, Lew AM, Wright MD, Heath WR, Shortman K, Lahoud MH (2008) The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood 112:3264–3273
Colonna M, Samaridis J, Angman L (2000) Molecular characterization of two novel C-type lectin-like receptors, one of which is selectively expressed in human dendritic cells. Eur J Immunol 30:697–704
Dissen E, Ryan JC, Seaman WE, Fossum S (1996) An autosomal dominant locus, Nka, mapping to the Ly-49 region of a rat natural killer (NK) gene complex, controls NK cell lysis of allogeneic lymphocytes. J Exp Med 183:2197–2207
Dissen E, Berg SF, Westgaard IH, Fossum S (1997) Molecular characterization of a gene in the rat homologous to human CD94. Eur J Immunol 27:2080–2086
Flornes LM, Bryceson YT, Spurkland A, Lorentzen JC, Dissen E, Fossum S (2004) Identification of lectin-like receptors expressed by antigen presenting cells and neutrophils and their mapping to a novel gene complex. Immunogenetics 56:506–517
Fuller GL, Williams JA, Tomlinson MG, Eble JA, Hanna SL, Pohlmann S, Suzuki-Inoue K, Ozaki Y, Watson SP, Pearce AC (2007) The C-type lectin receptors CLEC-2 and Dectin-1, but not DC-SIGN, signal via a novel YXXL-dependent signaling cascade. J Biol Chem 282:12397–12409
Hamann J, Montgomery KT, Lau S, Kucherlapati R, van Lier RA (1997) AICL: a new activation-induced antigen encoded by the human NK gene complex. Immunogenetics 45:295–300
Hao L, Klein J, Nei M (2006) Heterogeneous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc Natl Acad Sci USA 103:3192–3197
Hue S, Mention JJ, Monteiro RC, Zhang SL, Cellier C, Schmitz J, Verkarre V, Fodil N, Bahram S, Cerf-Bensussan N, Caillat-Zucman S (2004) A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity 21:367–377
Huysamen C, Brown GD (2009) The fungal pattern recognition receptor, Dectin-1, and the associated cluster of C-type lectin-like receptors. FEMS Microbiol Lett 290:121–128
Huysamen C, Willment JA, Dennehy KM, Brown GD (2008) CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. J Biol Chem 283:16693–16701
Kveberg L, Dai KZ, Westgaard IH, Daws MR, Fossum S, Naper C, Vaage JT (2009) Two major groups of rat NKR-P1 receptors can be distinguished based on chromosomal localization, phylogenetic analysis and Clr ligand binding. Eur J Immunol 39:541–551
Lorentzen JC, Flornes L, Eklow C, Backdahl L, Ribbhammar U, Guo JP, Smolnikova M, Dissen E, Seddighzadeh M, Brookes AJ, Alfredsson L, Klareskog L, Padyukov L, Fossum S (2007) Association of arthritis with a gene complex encoding C-type lectin-like receptors. Arthritis Rheum 56:2620–2632
Marshall AS, Willment JA, Lin HH, Williams DL, Gordon S, Brown GD (2004) Identification and characterization of a novel human myeloid inhibitory C-type lectin-like receptor (MICL) that is predominantly expressed on granulocytes and monocytes. J Biol Chem 279:14792–14802
Nagase M, Abe J, Takahashi K, Ando J, Hirose S, Fujita T (1998) Genomic organization and regulation of expression of the lectin-like oxidized low-density lipoprotein receptor (LOX-1) gene. J Biol Chem 273:33702–33707
Naper C, Hayashi S, Lovik G, Kveberg L, Niemi EC, Rolstad B, Dissen E, Ryan JC, Vaage JT (2002) Characterization of a novel killer cell lectin-like receptor (KLRH1) expressed by alloreactive rat NK cells. J Immunol 168:5147–5154
Nylenna O, Flornes LM, Westgaard IH, Woon PY, Naper C, Vaage JT, Gauguier D, Ryan JC, Dissen E, Fossum S (2005a) Killer cell lectin-like receptors and the natural killer cell gene complex. Glycobiology and Medicine 564:23–24
Nylenna O, Naper C, Vaage JT, Woon PY, Gauguier D, Dissen E, Ryan JC, Fossum S (2005b) The genes and gene organization of the Ly49 region of the rat natural killer cell gene complex. Eur J Immunol 35:261–272
Plougastel B, Dubbelde C, Yokoyama WM (2001) Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69. Immunogenetics 53:209–214
Pyz E, Huysamen C, Marshall AS, Gordon S, Taylor PR, Brown GD (2008) Characterisation of murine MICL (CLEC12A) and evidence for an endogenous ligand. Eur J Immunol 38:1157–1163
Renedo M, Arce I, Rodriguez A, Carretero M, Lanier LL, Lopez-Botet M, Fernandez-Ruiz E (1997) The human natural killer gene complex is located on chromosome 12p12–p13. Immunogenetics 46:307–311
Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL, Brown GD, Reis e Sousa C (2005) Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity 22:507–517
Saether PC, Westgaard IH, Flornes LM, Hoelsbrekken SE, Ryan JC, Fossum S, Dissen E (2005) Molecular cloning of KLRI1 and KLRI2, a novel pair of lectin-like natural killer-cell receptors with opposing signalling motifs. Immunogenetics 56:833–839
Sancho D, Gomez M, Sanchez-Madrid F (2005) CD69 is an immunoregulatory molecule induced following activation. Trends Immunol 26:136–140
Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M (2006) CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 440:540–544
Sobanov Y, Bernreiter A, Derdak S, Mechtcheriakova D, Schweighofer B, Duchler M, Kalthoff F, Hofer E (2001) A novel cluster of lectin-like receptor genes expressed in monocytic, dendritic and endothelial cells maps close to the NK receptor genes in the human NK gene complex. Eur J Immunol 31:3493–3503
Sun L, Zhao Y (2007) The biological role of dectin-1 in immune response. Int Rev Immunol 26:349–364
Suto Y, Yabe T, Maenaka K, Tokunaga K, Tadokoro K, Juji T (1997) The human natural killer gene complex (NKC) is located on chromosome 12p13.1-p13.2. Immunogenetics 46:159–162
Suzuki-Inoue K, Fuller GL, Garcia A, Eble JA, Pohlmann S, Inoue O, Gartner TK, Hughan SC, Pearce AC, Laing GD, Theakston RD, Schweighoffer E, Zitzmann N, Morita T, Tybulewicz VL, Ozaki Y, Watson SP (2006) A novel Syk-dependent mechanism of platelet activation by the C-type lectin receptor CLEC-2. Blood 107:542–549
Suzuki-Inoue K, Kato Y, Inoue O, Kaneko MK, Mishima K, Yatomi Y, Yamazaki Y, Narimatsu H, Ozaki Y (2007) Involvement of the snake toxin receptor CLEC-2, in podoplanin-mediated platelet activation, by cancer cells. J Biol Chem 282:25993–26001
Webb JR, Lee SH, Vidal SM (2002) Genetic control of innate immune responses against cytomegalovirus: MCMV meets its match. Genes Immun 3:250–262
Weis WI, Kahn R, Fourme R, Drickamer K, Hendrickson WA (1991) Structure of the calcium-dependent lectin domain from a rat mannose-binding protein determined by mad phasing. Science 254:1608–1615
Weis WI, Taylor ME, Drickamer K (1998) The C-type lectin superfamily in the immune system. Immunol Rev 163:19–34
Westgaard IH, Dissen E, Torgersen KM, Lazetic S, Lanier LL, Phillips JH, Fossum S (2003) the lectin-like receptor KLRE1 inhibits natural killer cell cytotoxicity. J Exp Med 197:1551–1561
Yokoyama WM, Ryan JC, Hunter JJ, Smith HR, Stark M, Seaman WE (1991) cDNA cloning of mouse NKR-P1 and genetic linkage with LY-49. Identification of a natural killer cell gene complex on mouse chromosome 6. J Immunol 147:3229–3236
Ziegler SF, Ramsdell F, Hjerrild KA, Armitage RJ, Grabstein KH, Hennen KB, Farrah T, Fanslow WC, Shevach EM, Alderson MR (1993) Molecular characterization of the early activation antigen CD69: a type II membrane glycoprotein related to a family of natural killer cell activation antigens. Eur J Immunol 23:1643–1648
Acknowledgments
This work was supported by The Norwegian Research Council, The Norwegian Cancer Society, and Bergljot and Sigurd Skaugen’s fund. The expert technical assistance of W. Jensen and M. Lauritzen is gratefully acknowledged. The authors have no conflicting financial interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 35 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Flornes, L.M., Nylenna, Ø., Saether, P.C. et al. The complete inventory of receptors encoded by the rat natural killer cell gene complex. Immunogenetics 62, 521–530 (2010). https://doi.org/10.1007/s00251-010-0455-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-010-0455-y