Abstract
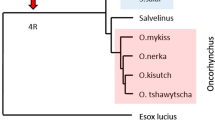
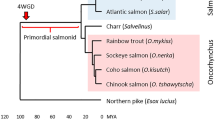
An unprecedented level of sequence diversity has been maintained in the salmonid major histocompatibility complex (MHC) class I UBA gene, with between lineage AA sequence identities as low as 34%. The derivation of deep allelic lineages may have occurred through interlocus exon shuffling or convergence of ancient loci with the UBA locus, but until recently, no such ancient loci were uncovered. Herein, we document the existence of eight additional MHC class I loci in salmon (UCA, UDA, UEA, UFA, UGA, UHA, ULA, and ZE), six of which share exon 2 and 3 lineages with UBA, and three of which have not been described elsewhere. Half of the UBA exon 2 lineages and all UBA exon 3 lineages are shared with other loci. Two loci, UGA and UEA, share only a single exon lineage with UBA, likely generated through exon shuffling. Based on sequence homologies, we hypothesize that most exchanges and duplications occurred before or during tetraploidization (50 to 100 Ma). Novel loci that share no relationship with other salmonid loci are also identified (UHA and ZE). Each locus is evaluated for its potential to function as a class Ia gene based on gene expression, conserved residues and polymorphism. UBA is the only locus that can indisputably be classified as a class Ia gene, although three of the eight loci (ZE, UCA, and ULA) conform in three out of four measures. We hypothesize that these additional loci are in varying states of degradation to class Ib genes.






Similar content being viewed by others
References
Aoyagi K, Dijkstra JM, Xia C, Denda I, Ototake M, Hashimoto K, Nakanishi T (2002) Classical MHC class I genes composed of highly divergent sequence lineages share a single locus in rainbow trout (Oncorhynchus mykiss). J Immunol 168:260–273
Beadle LC (1981) The inland waters of tropical Africa, 2nd edn. Longman, London
Bjorkman PJ, Parham P (1990) Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem 59:253–288
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Stominger JL, and Wiley DC (1987) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329:512–518
Carroll R (1988) Vertebrate palaeontology and evolution. Freeman, New York
Consuegra S, Megens H-J, Leon K, Stet RJM, Jordan WC (2005) Rapid evolution of the MH class I locus results in different allelic compositions in recently diverged populations of Atlantic salmon. Mol Biol Evol 22(4):1095–1106
Devlin RH (1993) Sequence of sockeye salmon type 1 and 2 growth hormone genes and the relationship of rainbow trout with Atlantic and Pacific salmon. Can J Fish Aquat Sci 50:1738–1748
Grimholt U, Drablos F, Jorgensen SM, Hoyheim B, Stet RJM (2002) The major histocompatibility class I locus in Atlantic salmon (Salmo salar L.): polymorphism, linkage analysis and protein modelling. Immunogenetics 54:570–581
Grimholt U, Larsen S, Nordmo R, Midtlyng P, Kjoeglum S, Storset A, Saebø S, Stet RJM (2003) MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics 55(4):210–219
Hansen JD, Strassburger P, Du Pasquier L (1996) Conservation of an α 2 domain within the teleostean world, MHC class I from the rainbow trout Oncorhynchus mykiss. Dev Comp Immunol 20:417–425
Kaufman J, Salomonsen J, Flajnik M (1994) Evolutionary conservation of MHC class I and class II molecules-different yet the same. Sem Immunol 6:411–424
Kiryu I, Dijkstra JM, Sarder RI, Fujiwara A, Yoshiura Y, Ototake M (2005) New MHC class Ia domain lineages in rainbow trout (Oncorhynchus mykiss) which are shared with other fish species. Fish Shellfish Immunol 18:243–254
Klein J (1991) Of HLA, tryps, and selection: an essay on coevolution of MHC and parasites. Hum Immunol 30:247–258
Klein D, Ono H, O’hUigin C, Vincek V, Goldschmidt T, Klein J (1993) Extensive MHC variability in cichlid fishes of Lake Malawi. Nature 364:330–334
Kruiswijk CP, Hermsen TT, Westphal AH, Savelkoul HFJ, Stet RJM (2002) A novel functional class I lineage in zebrafish (Danio rerio), carp (Cyprinus carpio), and large Barbus (Barbus intermedius) showing an unusual conservation of the peptide binding domains. J Immunol 169:1936–1947
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Miller KM, Withler RE (1996) Sequence analysis of a polymorphic Mhc class II gene in Pacific salmon. Immunogenetics 43:337–351
Miller KM, Withler RE (1997) MHC diversity in Pacific salmon: population structure and trans-species allelism. Hereditas 127:83–95
Miller KM, Withler RE (1998) The salmonid class I MHC: limited diversity in a primitive teleost. Immunol Rev 166:279–293
Miller KM, Ming TJ, Schulze AD, Wither RE (1999) Denaturing gradient gel electrophoresis (DGGE): a rapid and sensitive technique to screen nucleotide sequence variation in populations. Biotechniques 27(5):1017–1030
Miller KM, Kaukinen KH, Beacham TD, Withler RE (2001) Geographic heterogeneity in natural selection on an MHC locus in sockeye salmon. Genetica 111:237–257
Miller KM, Kaukinen KH, Schulze AD (2002) Expansion and contraction of major histocompatibility complex genes: a teleostean example. Immunogenetics 53:941–963
Myers RM, Fischer SG, Lerman LS, Maniatis T (1985) Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res 13:3131–3145
Persson AC, Stet RJM, Pilström L (1999) Characterization of MHC class I and B2-microglobulin sequences in Atlantic cod reveals an unusually high number of expressed class I genes. Immunogenetics 50:49–59
Phillips RB, Zimmerman A, Noakes MA, Palti Y, Morasch MRW, Eiben L, Ristow SS, Thorgaard GH, Hansen JD (2003) Physical and genetic mapping of the rainbow trout major histocompatibility regions: evidence for duplication of the class I region. Immunogenetics 55:561–569
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): population genetics software for exact tests and ecumenism. Heredity 86:248–249
Richman A (2000) Evolution of balanced genetic polymorphism. Mol Ecol 9:1952–1963
Saper MA, Bjorkman PJ, Wiley DE (1991) Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 Å resolution. J Mol Biol 219:277–319
Shiina T, Dijkstra JM, Shimizu S, Watanabe A, Yanagiya K, Kiryu I, Fujiwara A, Nishida-Umehara C, Kaba Y, Hirono I, Yoshiura Y, Aoki T, Inoko H, Kulski JK, Ototake M (2005) Interchromosomal duplication of major histocompatibility complex class I regions in rainbow trout (Oncorhynchus mykiss), a species with a presumably recent tetraploid ancestry. Immunogenetics 56:878–893
Shum BP, Rajalingam R, Magor KE, Azumi K, Carr WH, Dixon B, Stet RJ, Adkison MA, Hedrick RP, Parham P (1999) A divergent non-classical class I gene conserved in salmonids. Immunogenetics 49(6):479–490
Shum BP, Guethlein L, Flodin LR, Adkison MA, Hedrick RP, Nehring RB, Stet RJM, Secombes C, Parham P (2001) Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J Immunol 166:3297–3308
Shum BP, Mason PM, Magor KE, Flodin LR, Stet RJM, Parham P (2002) Structures of two major histocompatibility complex (MHC) class I genes of the Rainbow trout (Oncorhynchus mykiss). Immunogenetics 54(3):193–199
Stet RJ, Kruiswijk CP, Dixon B (2003) Major histocompatibility lineages and immune gene function in teleost fishes: the road not taken. Crit Rev Immunol 23(5–6):441–471, Review
Takahata N (1995) MHC diversity and selection. Immunol Rev 143:225–247
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Young WP, Wheeler PA, Fields RD, Thorgaard GH (1996) DNA fingerprinting confirms isogenicity of androgenetically-derived rainbow trout lines. J Hered 87:77–81
Acknowledgements
We thank Gary Thorgaard (Washington State University) who kindly provided us DNA/RNA tissues from clonal lines of rainbow trout and Heritage Seafarms who supplied the Atlantic salmon families. IHNV infected fish were provided by Garth Traxler and additional Atlantic salmon samples were provided by Robert Devlin, both of Fisheries & Oceans Canada. Janine Supernault, Norma Ginther, and Liane Stenhouse assisted with sampling and DNA/RNA extractions. Ruth Withler and the three anonymous reviewers are also thanked for their input on revisions of the manuscript. This work was supported by CBS Genomics Research and Development Program Fund.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Miller, K.M., Li, S., Ming, T.J. et al. The salmonid MHC class I: more ancient loci uncovered. Immunogenetics 58, 571–589 (2006). https://doi.org/10.1007/s00251-006-0125-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-006-0125-2