Abstract
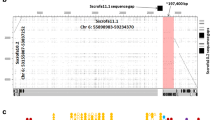
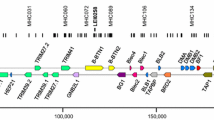
Continuous genomic sequence has been previously determined for the swine leukocyte antigen (SLA) class I region from the TNF gene cluster at the border between the major histocompatibility complex (MHC) class III and class I regions to the UBD gene at the telomeric end of the classical class I gene cluster (SLA-1 to SLA-5, SLA-9, SLA-11). To complete the genomic sequence of the entire SLA class I genomic region, we have analyzed the genomic sequences of two BAC clones carrying a continuous 237,633-bp-long segment spanning from the TRIM15 gene to the UBD gene located on the telomeric side of the classical SLA class I gene cluster. Fifteen non-class I genes, including the zinc finger and the tripartite motif (TRIM) ring-finger-related family genes and olfactory receptor genes, were identified in the 238-kilobase (kb) segment, and their location in the segment was similar to their apparent human homologs. In contrast, a human segment (alpha block) spanning about 375 kb from the gene ETF1P1 and from the HLA-J to HLA-F genes was absent from the 238-kb swine segment. We conclude that the gene organization of the MHC non-class I genes located in the telomeric side of the classical SLA class I gene cluster is remarkably similar between the swine and the human segments, although the swine lacks a 375-kb segment corresponding to the human alpha block.




Similar content being viewed by others
References
Dummy
Amadou C (1999) Evolution of the Mhc class I region: the framework hypothesis. Immunogenetics 49:362–367
Amadou C, Younger RM, Sims S, Mathews LH, Rogers J, Kumanovics A, Ziegler A, Beck S, Lindahl KF (2003) Co-duplication of olfactory receptor and MHC class I genes in the mouse major histocompatibility complex. Hum Mol Genet 12:3025–3040
Anzai T, Shiina T, Kimura N, Yanagiya K, Kohara S, Shigenari A, Yamagata T, Kulski JK, Naruse TK, Fujimori Y, Fukuzumi Y, Yamazaki M, Tashiro H, Iwamoto C, Umehara Y, Imanishi T, Meyer A, Ikeo K, Gojobori T, Bahram S, Inoko H (2003) Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc Natl Acad Sci U S A 100:7708–7713
Beck TW, Menninger J, Murphy WJ, Nash WG, O’Brien SJ, Yuhki N (2005) The feline major histocompatibility complex is rearranged by an inversion with a breakpoint in the distal class I region. Immunogenetics 56:702–709
Bernardi G (1993) The isochore organization of the human genome and its evolutionary history—a review. Gene 135:57–66
Chardon P, Renard C, Vaiman M (1999a) The major histocompatibility complex in swine. Immunol Rev 167:179–192
Chardon P, Rogel-Gaillard C, Peelman L, Yerle M, Verten FW, Renard C, Vaiman M (1999b) Physical organization of the pig major histocompatibility complex class II region. Immunogenetics 50:344–348
Chardon P, Renard C, Rogel-Gaillard C, Vaiman M (2000) The porcine major histocompatibility complex and related paralogous regions: a review. Genet Sel Evol 32:109–128
Chardon P, Rogel-Gaillard C, Cattolico L, Duprat S, Vaiman M, Renard C (2001) Sequence of the pig major histocompatibility region containing all non-classical class I genes. Tissue Antigens 57:55–65
Deininger PL (1983) Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem 129:216–223
Gustafson AL, Tallmadge RL, Ramlachan N, Miller D, Bird H, Antcza DF (2003) An ordered BAC contig map of the equine major histocompatibility complex. Cytogenet Genome Res 102:189–195
Havlickova M, Prezeau L, Duthey B, Bettler B, Pin JP, Blahos J (2002) The intracellular loops of the GB2 subunit are crucial for G-protein coupling of the heteromeric gamma-aminobutyrate B receptor. Mol Pharmacol 62:343–350
Hurt P, Walter L, Sudbrak R, Klages S, Muller I, Shiina T, Inoko H, Lehrach H, Gunther E, Reinhardt R, Himmelbauer H (2004) The genomic sequence and comparative analysis of the rat major histocompatibility complex. Genome Res 14:631–639
Kaupmann K, Huggel K, Heid J, Flor PJ, Bischoff S, Mickel SJ, MacMaster G, Ansdt C, Bittiger H, Froestl W et al (1997) Expression cloning of GABA(B) receptors uncovers similarity to metabotropic glutamate receptors. Nature 386:239–246
Kostia S, Ruohonen-Lehto M, Vainola R, Varvio SL (2000) Phylogenetic information in inter-SINE and inter-SSR fingerprints of the artiodactyla and evolution of the bov-tA SINE. Heredity 84:37–45
Kulski JK, Gaudieri S, Dawkins RL (2000) Using Alu J elements as molecular clocks to trace the evolutionary relationships between duplicated HLA class I segments. J Mol Evol 50:510–519
Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H (2002) Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev 190:95–122
Kulski JK, Anzai T, Shiina T, Inoko H (2004) MHC class I duplicon structures, organization and evolution within the alpha block of the rhesus macaque. Mol Biol Evol 21:2079–2091
Kumanovics A, Takada T, Lindahl KF (2003) Genomic organization of the mammalian MHC. Annu Rev Immunol 21:629–657
Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN, Murphy CN, Carter DB, Hawley RJ, Prather RS (2002) Production of alpha-1,3-galactosyltaransferase knockout pigs by nuclear transfer cloning. Science 295:1089–1092
Meyer M, Gaudieri S, Rhodes DA, Trowsdale J (2003) Cluster of TRIM genes in the human MHC class I region sharing the B30.2 domain. Tissue Antigens 61:63–71
Mizuki N, Ando H, Kimura M, Ohno S, Miyata S, Yamazaki M, Tashiro H, Watanabe K, Ono A, Taguchi S, Sugawara C, Fukuzumi Y, Okumura K, Goto K, Ishihara M, Nakamura S, Yonemoto J, Kikuti YY, Shiina T, Chen L, Ando A, Ikemura T, Inoko H (1997) Nucleotide sequence analysis of the HLA class I region spanning the 237-kb segment around the HLA-B and -C genes. Genomics 42:55–66
Mungall AJ, Palmer SA, Sims SK, Edwards CA et al (2003) The DNA sequence and analysis of human chromosome 6. Nature 425:805–811
Peelman LJ, Chardon P, Vaiman M, Mattheeuws M, Van Zeveren A, Van de Weghe A, Bouquet Y, Campbell RD (1996) A detailed physical map of the porcine major histocompatibility complex (MHC) class III region: comparison with human and mouse MHC class III regions. Mamm Genome 7:363–367
Rabin M, Fries R, Singer DS, Ruddle FH (1985) Assignment of the porcine major histocompatibility complex to chromosome 7 by in situ hybridization. Cytogenet Cell Genet 39:206–209
Renard C, Vaiman M, Chiannikulchai N, Cattolico L, Robert C, Chardon P (2001) Sequence of the pig major histocompatibility region containing the classical class I genes. Immunogenetics 53:490–500
Renard C, Chardon P, Vaiman M (2003) The phylogenetic history of the MHC class I gene families in pig, including a fossil gene predating mammalian radiation. J Mol Evol 57:420–434
Rogel-Gaillard C, Bourgeaux N, Billault A, Vaiman M, Chardon P (1999) Construction of a swine BAC library: application to the characterization and mapping of porcine type C endoviral elements. Cytogenet Cell Genet 85:205–211
Sachs DH (1994) The pig as a potential xenograft donor. Vet Immunol Immunopathol 43:185–191
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10: 577–586
Shigenari A, Ando A, Renard C, Chardon P, Shiina T, Kulski JK, Yasue H, Inoko H (2004) Nucleotide sequencing analysis of the swine 433-kb genomic segment located between the non-classical and classical SLA class I gene clusters. Immunogenetics 55:695–705
Shiina T, Tamiya G, Oka A, Yamagata T, Yamagata N, Kikkawa E, Goto K, Mizuki N, Watanabe K, Fukuzumi Y, Taguchi S, Sugawara C, Ono A, Chen L, Yamazaki M, Tashiro H, Ando A, Ikemura T, Kimura M, Inoko H (1998) Nucleotide sequencing analysis of the 146 kb segment around the IkBL and MICA genes at the centromeric end of the HLA class I region. Genomics 47:372–382
Shiina T, Inoko H, Kulski JK (2004) An update of the HLA genomic region, locus information and disease association: 2004. Tissue Antigens 64:631–649
Smith TP, Rohrer GA, Alexander LJ, Troyer DL, Kirby-Dobbels KR, Janzen MA, Cornwell DL, Louis CF, Schook, LB, Beattie CW (1995) Directed integration of the physical and genetic linkage maps of the swine chromosome 7 reveals that SLA spans the centromere. Genome Res 5:259–271
Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, Humphray SJ, Hunt S, Mungall AJ, Osoegawa K, Palmer S, Roberts AN, Rogers J, Sims S, Wang Y, Wilming LG, Elliott JF, de Jong PJ, Sawcer S, Todd JA, Trowsdale J, Beck S (2004) Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 14:1176–1187
The MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex (MHC). Nature 401:921–923
Velten F, Rogel-Gaillard C, Renard C, Pontarotti P, Tazi-Ahnini R, Vaiman M, Chardon P (1998) A first map of the porcine major histocompatibility complex class I region. Tissue Antigens 51:183–194
Velten F, Renald C, Rogel-Gaillard C, Vaiman M, Schrezenmeir J, Chardon P (1999) Spatial arrangement of pig MHC class I sequences. Immunogenetics 49:919–930
Weissert R, Kuhle J, de Graaf KL, Wienhold W, Herrmann MM, Muller C, Forsthuber TG, Wiesmuller KH, Melms A (2002) High immunogenicity of intracellular myelin oligodendrocyte glycoprotein epitopes. J Immunol 169:548–556
Xu Y, Rothchild MF, Warmer CM (1992) Mapping of the SLA complex of the miniature swine mapping of the SLA gene complex by pulsed field gel electrophoresis. Mamm Genome 2:2–10
Younger RM, Amadou C, Bethel G, Ehlers A, Lindahl KF, Forbes S, Horton R, Milne S, Mungall AJ, Trowsdal F, Volz A, Ziegler A, Beck S (2001) Characterization of clustered MHC-linked olfactory receptor genes in human and mouse. Genome Res 11:519–530
Zhang X, Firestein S (2002) The olfactory receptor gene superfamily of the mouse. Nat Neurosci 5:124–133
Zhang J, Zhang L, Zhao S, Lee EY (1998) Identification and characterization of the human HCG V gene product as a novel inhibitor of protein phosphatase-1. Biochemistry 37:16728–16734
Website References
http://www.repeatmasker.org/; RepeatMasker program.
http://www.ncbi.nlm.nih.gov:80/BLAST/; BLAST search programs.
http://www.ebi.ac.uk/fasta33/; BLAST search programs.
http://genes.mit.edu/GENSCAN.html; GENSCAN search programs.
http://pipmaker.bx.psu.edu/pipmaker/; PipMaker programs.
http://bioinformatics.weizmann.ac.il/cards/; Human genes database.
http://www.ncbi.nlm.nih.gov/; Site for Map Viewer and GenBank database.
http://www.ncbi.nlm.nih.gov/ LocusLink/; LocusLink database.
http://www.sanger.ac.uk/HGP/Chr6/; Sanger Institute human MHC sequence.
Acknowledgements
This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Animal Genome Research Project of the Ministry of Agriculture, Forestry and Fisheries of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported in this paper have been submitted to DDBJ, EMBL, and GenBank databases under accession numbers AB158486 and AB158487
Rights and permissions
About this article
Cite this article
Ando, A., Shigenari, A., Kulski, J.K. et al. Genomic sequence analysis of the 238-kb swine segment with a cluster of TRIM and olfactory receptor genes located, but with no class I genes, at the distal end of the SLA class I region. Immunogenetics 57, 864–873 (2005). https://doi.org/10.1007/s00251-005-0053-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-005-0053-6