Abstract
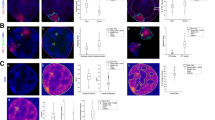
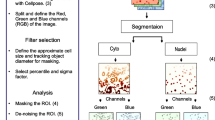
Data segmentation and object rendering is required for localization super-resolution microscopy, fluorescent photoactivation localization microscopy (FPALM), and direct stochastic optical reconstruction microscopy (dSTORM). We developed and validated methods for segmenting objects based on Delaunay triangulation in 3D space, followed by facet culling. We applied them to visualize mitochondrial nucleoids, which confine DNA in complexes with mitochondrial (mt) transcription factor A (TFAM) and gene expression machinery proteins, such as mt single-stranded-DNA-binding protein (mtSSB). Eos2-conjugated TFAM visualized nucleoids in HepG2 cells, which was compared with dSTORM 3D-immunocytochemistry of TFAM, mtSSB, or DNA. The localized fluorophores of FPALM/dSTORM data were segmented using Delaunay triangulation into polyhedron models and by principal component analysis (PCA) into general PCA ellipsoids. The PCA ellipsoids were normalized to the smoothed volume of polyhedrons or by the net unsmoothed Delaunay volume and remodeled into rotational ellipsoids to obtain models, termed DVRE. The most frequent size of ellipsoid nucleoid model imaged via TFAM was 35 × 45 × 95 nm; or 35 × 45 × 75 nm for mtDNA cores; and 25 × 45 × 100 nm for nucleoids imaged via mtSSB. Nucleoids encompassed different point density and wide size ranges, speculatively due to different activity stemming from different TFAM/mtDNA stoichiometry/density. Considering twofold lower axial vs. lateral resolution, only bulky DVRE models with an aspect ratio >3 and tilted toward the xy-plane were considered as two proximal nucleoids, suspicious occurring after division following mtDNA replication. The existence of proximal nucleoids in mtDNA-dSTORM 3D images of mtDNA “doubling”-supported possible direct observations of mt nucleoid division after mtDNA replication.









Similar content being viewed by others
References
Abdi H, Williams LJ (2010) Principal component analysis. Wiley Interdiscip Rev Computat Stat 2:433–459
Antonicka H, Sasarman F, Nishimura T, Paupe V, Shoubridge EA (2013) The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab 17:386–398
Aquino D, Schonle A, Geisler C, Middendorff CV, Wurm CA, Okamura Y, Lang T, Hell SW, Egner A (2011) Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores. Nat Methods 8:353–359
Asin-Cayuela J, Gustafsson CM (2007) Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem Sci 32:111–117
Baddeley D, Jayasinghe ID, Lam L, Rossberger S, Cannell MB, Soeller C (2009) Optical single-channel resolution imaging of the ryanodine receptor distribution in rat cardiac myocytes. Proc Natl Acad Sci USA 106:22275–22280
Blomain ES, McMahon SB (2012) Dynamic regulation of mitochondrial transcription as a mechanism of cellular adaptation. Biochim Biophys Acta 1819:1075–1079
Bogenhagen DF (2012) Mitochondrial DNA nucleoid structure. Biochim Biophys Acta 1819:914–920
Bogenhagen DF, Rousseau D, Burke S (2008) The layered structure of human mitochondrial DNA nucleoids. J Biol Chem 283:3665–3675
Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA (2011) Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol Cell Biol 31:4994–5010
Cameron JM, Levandovskiy V, Mackay N, Ackerley C, Chitayat D, Raiman J, Halliday WH, Schulze A, Robinson BH (2011) Complex V TMEM70 deficiency results in mitochondrial nucleoid disorganization. Mitochondrion 11:191–199
Campbell CT, Kolesar JE, Kaufman BA (2012) Mitochondrial transcription factor A regulates mitochondrial transcription initiation. DNA packaging, and genome copy number. Biochim Biophys Acta 1819:921–929
Delaunay B (1934) Sur la sphère vide, Izvestia Akademii Nauk SSSR, Otdelenie Matematicheskikh i Estestvennykh Nauk 7:793–800
Dlasková A, Špaček T, Šantorová J, Plecitá-Hlavatá L, Berková Z, Saudek F, Lessard M, Bewersdorf J, Ježek P (2010) 4Pi microscopy reveals an impaired three-dimensional mitochondrial network of pancreatic islet beta-cells in an experimental model of type-2 diabetes. Biochim Biophys Acta Bioenerg 1797:1327–1341
Dlasková A, Engstová H, Plecitá-Hlavatá L, Lessard M, Alán L, Reguera DP, Jabůrek M, Ježek P (2015) Distribution of mitochondrial DNA nucleoids inside the linear tubules vs. bulk parts of mitochondrial network as visualized by 4Pi microscopy. J Bioenerg Biomembr 47:255–263
Elachouri G, Vidoni S, Zanna C, Pattyn A, Boukhaddaoui H, Gaget K, Yu-Wai-Man P, Gasparre G, Sarzi E, Delettre C, Olichon A, Loiseau D, Reynier P, Chinnery PF, Rotig A, Carelli V, Hamel CP, Rugolo M, Lenaers G (2011) OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res 21:12–20
Falkenberg M, Larsson NG, Gustafsson CM (2007) DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem 76:679–699
Gauthier BR, Wiederkehr A, Baquié M, Dai C, Powers AC, Kerr-Conte J, Pattou F, MacDonald RJ, Ferrer J, Wollheim CB (2009) PDX1 deficiency causes mitochondrial dysfunction and defective insulin secretion through TFAM suppression. Cell Metab 10:110–118
Gilkerson RW, Schon EA, Hernandez E, Davidson MM (2008) Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol 181:1117–1128
Gould TJ, Burke D, Bewersdorf J, Booth MJ (2012) Adaptive optics enables 3D STED microscopy in aberrating specimens. Opt Express 20:20998–21009
Han KY, Ha T (2015) Dual-color three-dimensional STED microscopy with a single high-repetition-rate laser. Opt Lett 40:2653–2656
Hoke GD, Pavco PA, Ledwith BJ, Van Tuyle GC (1990) Structural and functional studies of the rat mitochondrial single strand DNA binding protein P16. Arch Biochem Biophys 282:116–124
Holt IJ (2009) Mitochondrial DNA replication and repair: all a flap. Trends Biochem Sci 34:358–365
Holt IJ (2010) Zen and the art of mitochondrial DNA maintenance. Trends Genet 26:103–109
Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN (2007) Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion 7:311–321
Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, Bennett BT, Hess ST, Bewersdorf J (2008) 3D sub-100 nm resolution by biplane fluorescence photoactivation localization microscopy. Nat Methods 5:527–529
Kaguni LS (2004) DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem 73:293–320
Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF (2012) Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc Natl Acad Sci USA 109:6136–6141
Korhonen JA, Gaspari M, Falkenberg M (2003) TWINKLE Has 5′–>3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem 278:48627–48632
Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jacobs S (2011) Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci USA 108:13534–13539
Kukat C, Davies KM, Wurm CA, Spåhr H, Bonekamp NA, Kühl I, Joos F, Polosa PL, Park CB, Posse V, Falkenberg M, Jakobs S, Kühlbrandt W, Larsson NG (2015) Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci USA 112:11288–11293
Lee KW, Okot-Kotber C, LaComb JF, Bogenhagen DF (2013) Mitochondrial ribosomal RNA (rRNA) methyltransferase family members are positioned to modify nascent rRNA in foci near the mitochondrial DNA nucleoid. J Biol Chem 288:31386–31399
Milenkovic D, Matic S, Kühl I, Ruzzenente B, Freyer C, Jemt E, Park CB, Falkenberg M, Larsson NG (2013) TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum Mol Genet 22:1983–1993
Mlodzianoski MJ, Schreiner JM, Callahan SP, Smolková K, Dlasková A, Šantorová J, Ježek P, Bewersdorf J (2011) Sample drift correction in 3D fluorescence photoactivation localization microscopy. Opt Express 19:15009–15019
Ngo HB, Kaiser JT, Chan DC (2011) The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol 18:1290–1296
Osseforth C, Moffitt JR, Schermelleh L, Michaelis J (2014) Simultaneous dual-color 3D STED microscopy. Opt Express 22:7028–7039
Plecitá-Hlavatá L, Lessard M, Šantorová J, Bewersdorf J, Ježek P (2008) Mitochondrial oxidative phosphorylation and energetic status are reflected by morphology of mitochondrial network in INS-1E and HEP-G2 cells viewed by 4Pi microscopy. Biochim Biophys Acta 1777:834–846
Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA (2004) Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 64:985–993
Ruhanen H, Borrie S, Szabadkai G, Tyynismaa HH, Jones AW, Kang D, Taanman JW, Yasukawa T (2010) Mitochondrial single-stranded DNA binding protein is required for maintenance of mitochondrial DNA and 7S DNA but is not required for mitochondrial nucleoid organization. Biochim Biophys Acta 1803:931–939
Scarpulla RC (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88:611–638
Schmidt R, Wurm CA, Punge A, Egner A, Jakobs S, Hell SW (2009) Mitochondrial cristae revealed with focused light. Nano Lett 9:2508–2510
Shi CM, Xu GF, Yang L, Fu ZY, Chen L, Fu HL, Shen YH, Zhu L, Ji CB, Guo XR (2013) Overexpression of TFAM Protects 3T3-L1 Adipocytes from NYGGF4 (PID1) overexpression-induced insulin resistance and mitochondrial dysfunction. Cell Biochem Biophys 66:489–497
Spelbrink JN (2010) Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life 62:19–32
Tauber J, Dlasková A, Šantorová J, Smolková K, Alán L, Špaček T, Plecitá-Hlavatá L, Jabůrek M, Ježek P (2013) Distribution of mitochondrial nucleoids upon mitochondrial network fragmentation and network reintegration in HEPG2 cells. Int J Biochem Cell Biol 45:593–603
Van Tuyle GC, Pavco PA (1985) The rat liver mitochondrial DNA-protein complex: displaced single strands of replicative intermediates are protein coated. J Cell Biol 100:251–257
Wildanger D, Medda R, Kastrup L, Hell SW (2009) A compact STED microscope providing 3D nanoscale resolution. J Microsc 236:35–43
Acknowledgments
The authors thank Martin Bartoš (Alef, Ltd., Prague) for help with nucleoid modeling using the Delaunay algorithm and Paraview software; and to Prof. Daniel F. Bogenhagen (Department of Pharmacological Sciences, State University of New York at Stony Brook) for providing the Eos2 vector and rabbit anti-TFAM antibodies. The project was principally supported by a grant of the Grant Agency of the Czech Republic (GACR) No. 13-02033S to P.J.; by the research project RVO67985823 to the Institute of Physiology; and also by the project BIOCEV—Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University (CZ.1.05/1.1.00/02.0109), from the European Regional Development Fund. The latter source was also co-funded by the European Social Fund and the state budget of the Czech Republic.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Alán, L., Špaček, T. & Ježek, P. Delaunay algorithm and principal component analysis for 3D visualization of mitochondrial DNA nucleoids by Biplane FPALM/dSTORM. Eur Biophys J 45, 443–461 (2016). https://doi.org/10.1007/s00249-016-1114-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-016-1114-5