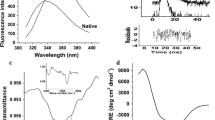
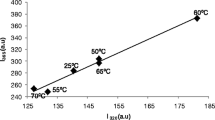
Abstract
Because of their vast diversity of substrate specificity and reaction conditions, lipases are versatile materials for biocatalysis. Lipase A from Bacillus subtilis (BSLA) is the smallest lipase yet discovered. It has the typical α/β hydrolase fold but lacks a lid covering the substrate cleft. In this study, the pH-dependence of the activity, stability, structure, and dynamics of BSLA was investigated by fluorescence spectroscopy. By use of a fluorogenic substrate it was revealed that the optimum pH for BSLA activity is 8.5 whereas thermodynamic and kinetic stability are maximum at pH 10. The origin of this behavior was clarified by investigation of ANS (8-anilino-1-naphthalenesulfonic acid) binding and fluorescence quenching of the two single tryptophan mutants W31F and W42F. Variations in segmental dynamics were investigated by use of time-resolved fluorescence anisotropy. This analysis showed that the activity maximum is governed by high surface hydrophobicity and high segmental mobility of surface loops whereas the stability optimum is a result of low segmental mobility and surface hydrophobicity.









Similar content being viewed by others
References
Acharya P, Rao NM (2003) Stability studies on a lipase from Bacillus subtilis in guanidinium chloride. J Protein chem 22(1):51–60
Ahmad S, Kamal MZ, Sankaranarayanan R, Rao NM (2008) Thermostable Bacillus subtilis lipases. In vitro evolution and structural insight. J Mol Biol 381(2):324–340
Anunciado D, Agumeh M, Kormos BL, Beveridge DL, Knee JL, Baranger AM (2008) Characterization of the dynamics of an essential helix in the U1A protein by time-resolved fluorescence measurements. J Phys Chem B 112(19):6122–6130
Augustyniak W, Brzezinska AA, Pijning T, Wienk H, Boelens R, Dijkstra BW, Reetz MT (2012) Biophysical characterization of mutants of Bacillus subtilis lipase evolved for thermostability: factors contributing to increased activity retention. Protein Sci 21(4):487–497
Austin RH, Beeson KW, Eisenstein L, Frauenfelder H, Gunsalus IC (1975) Dynamics of ligand binding to myoglobin. Biochemistry 14(24):5355–5373
Beechem JM, Brand L (1985) Time-resolved fluorescence of proteins. Annu Rev Biochem 54(1):43–71
Bornscheuer UT, Bessler C, Srinivas R, Hari Krishna S (2002) Optimizing lipases and related enzymes for efficient application. Trends Biotechnol 20(10):433–437
Brzozowski AM, Savage H, Verma CS, Turkenburg JP, Lawson DM, Svendsen A, Patkar S (2000) Structural origins of the interfacial activation in thermomyces (Humicola) lanuginosa lipase. Biochemistry 39(49):15071–15082
Chahinian H, Snabe T, Attias C, Fojan P, Petersen SB, Carrière F (2006) How gastric lipase, an interfacial enzyme with a Ser-His-Asp catalytic triad, acts optimally at acidic pH. Biochemistry 45(3):993–1001
Chakraborty S, Ittah V, Bai P, Luo L, Haas E, Peng Z-Y (2001) Structure and dynamics of the α-lactalbumin molten globule: fluorescence studies using proteins containing a single tryptophan residue †. Biochemistry 40(24):7228–7238
Dusa A, Kaylor J, Edridge S, Bodner N, Hong D-P, Fink AL (2006) Characterization of oligomers during alpha-synuclein aggregation using intrinsic tryptophan fluorescence. Biochemistry 45(8):2752–2760
Eggert T, Leggewie C, Puls M, Streit W, van Pouderoyen G, Dijkstra BW, Jaeger K-E (2004) Novel biocatalysts by identification and design. Biocatal Biotransformat 22(2):141–146
Greene RF, Pace CN (1974) Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, α-chymotrypsin, and β-lactoglobulin. J Biol Chem 249(17):5388–5393
Henzler-Wildman K, Kern D (2007) Dynamic personalities of proteins. Nature 450(7172):964–972. doi:10.1038/nature06522
Jaeger K-E, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13(4):390–397
Kamal MZ, Ahmad S, Molugu TR, Vijayalakshmi A, Deshmukh MV, Sankaranarayanan R, Rao NM (2011) In vitro evolved non-aggregating and thermostable lipase: structural and thermodynamic investigation. J Mol Biol 413(3):726–741
Kamal MZ, Mohammad TAS, Krishnamoorthy G, Rao NM (2012) Role of active site rigidity in activity: MD simulation and fluorescence study on a lipase mutant. PLoS One 7(4):e35188
Kawasaki K, Kondo H, Suzuki M, Ohgiyai S, Tsuda S (2002) Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1.3 a resolution. Acta Crystallogr D Biol Crystallogr 58(7):1168–1174
Kinosita K, Kawato S, Ikegami A (1977) A theory of fluorescence polarization decay in membranes. Biophys J 20(3):289–305
Lakowicz JR (2006) Principles of fluorescence spectroscopy. Springer, LLC, Boston
Lesuisse E, Schanck K, Colson C (1993) Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur J Biochem 216(1):155–160
Manafi M, Kneifel W, Bascomb S (1991) Fluorogenic and chromogenic substrates used in bacterial diagnostics. Microbiol Rev 55(3):335–348
Newman J (2004) Novel buffer systems for macromolecular crystallization. Acta crystallogr Sect D Biol Crystallogr 60(Pt 3):610–612
Pace CN, Grimsley GR, Scholtz JM (2008) Denaturation of proteins by urea and guanidine hydrochloride. In: Fersht AR (ed) Protein science encyclopedia. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim
Pencreac’h G, Graille J, Pina M, Verger R (2002) An ultraviolet spectrophotometric assay for measuring lipase activity using long-chain triacyglycerols from aleurites fordii seeds. Anal Biochem 303(1):17–24
Petersen MTN, Fojan P, Petersen SB (2001) How do lipases and esterases work: the electrostatic contribution. J Biotechnol 85(2):115–147
Pettit FK, Bare E, Tsai A, Bowie JU (2007) HotPatch: a statistical approach to finding biologically relevant features on protein surfaces. J Mol Biol 369(3):863–879
Rajakumara E, Acharya P, Ahmad S, Sankaranaryanan R, Rao NM (2008) Structural basis for the remarkable stability of Bacillus subtilis lipase (Lip A) at low pH. Biochim et Biophys Acta Proteins Proteomics 1784(2):302–311
Reetz MT (2002) Lipases as practical biocatalysts. Curr Opin Chem Biol 6(2):145–150
Rosen CG, Weber G (1969) Dimer formation from 1-anilino-8-naphthalenesulfonate catalyzed by bovine serum albumin. Fluorescent molecule with exceptional binding properties. Biochemistry 8(10):3915–3920
Santoro MM, Bolen DW (1988) Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl.alpha.-chymotrypsin using different denaturants. Biochemistry 27(21):8063–8068
Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409(6817):258–268
Schröder GF, Alexiev U, Grubmüller H (2005) Simulation of fluorescence anisotropy experiments: probing protein dynamics. Biophys J 89(6):3757–3770
Szabo A (1984) Theory of fluorescence depolarization in macromolecules and membranes. J Chem Phys 81(1):150
van Pouderoyen G, Eggert T, Jaeger K-E, Dijkstra BW (2001) The crystal structure of Bacillus subtili lipase: a minimal α/β hydrolase fold enzyme. J Mol Biol 309(1):215–226
Yu Y, Wu D, Liu C, Zhao Z, Yang Y, Li Q (2012) Lipase/esterase-catalyzed synthesis of aliphatic polyesters via polycondensation: a review. Process Biochem 47(7):1027–1036
Acknowledgments
We thank Dr Ulrich Kraus, Forschungszentrum Jülich, for kindly providing the DNA of BSLA and the purification procedure, and Professor Nickisch-Hartfiel for making it possible to conduct parts of this research in the Biotechnology Laboratories of the Chemistry Department.
Funding sources
The work was supported by the “Professorinnenprogramm des Bundes und der Länder”, 01FP09231B.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kübler, D., Ingenbosch, K.N., Bergmann, A. et al. Fluorescence spectroscopic analysis of the structure and dynamics of Bacillus subtilis lipase A governing its activity profile under alkaline conditions. Eur Biophys J 44, 655–665 (2015). https://doi.org/10.1007/s00249-015-1061-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-015-1061-6